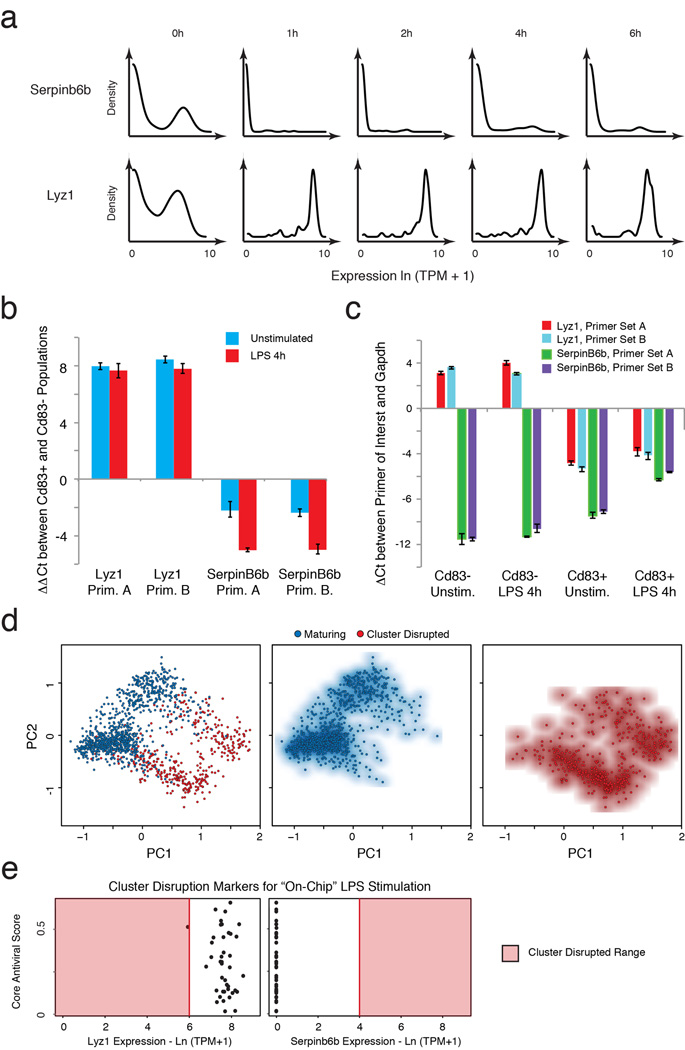
Extended Figure 4. Cluster disruption.
(a) Single-cell expression distributions for Serpinb6b (a positive marker of cluster disruption) and Lyz1 (a negative marker of cluster disruption) at each time point (marked on top) after stimulation with LPS (all cells included, see SI). Distributions are scaled to have the same maximum height. (b) Difference in mRNA expression as measured by qRT-PCR (with a Gapdh control) between Lyz1 or SerpinB6b in cells pre-sorted before stimulation to express or not express CD83 (CD83+ and CD83−, respectively), a known cell surface marker of cluster-disrupted cells (see SI). Pre-sorted cells were then either unstimulated (blue) or stimulated (red) with LPS for 4h. (c) Expression of cluster-disruption markers does not change with stimulation. qRT-PCR showing the difference between Gapdh (control) and Lyz1 or SerpinB6b expression in cells pre-sorted on Cd83 either in the presence or absence of simulation with LPS. (d) PCA showing the separation between “maturing” (blue dots) and “cluster-disrupted” (red dots) cells. (e) Expression of cluster disruption markers of cells stimulated with LPS “on chip”. For each cell (black dot) stimulated with LPS “on chip”, shown are the expression levels (X axis) of Serpin6b (cluster disruption cell marker, left) and Lyz1 (normal maturing cell marker, right) vs. its antiviral score (Y axis). With one exception, the cells are clearly “maturing” and not “cluster-disrupted”. Red shading: range of expression in cluster disrupted cells.