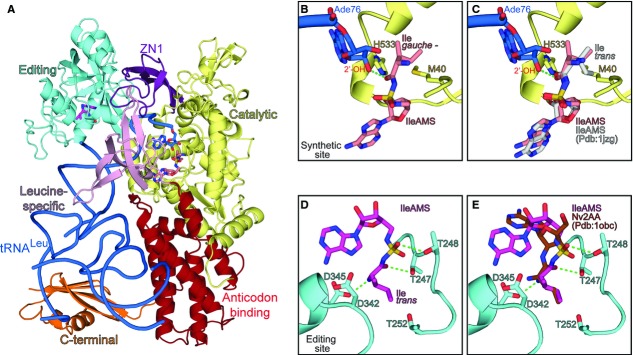
Cartoon representation of the EcLeuRS:tRNALeu:Ile-AMS ternary complex showing the different domains colored as follows: yellow, catalytic domain; purple, ZN1 domain; cyan, editing domain; pink, leucine-specific domain; red, anticodon-binding domain; orange, C-terminal domain. The tRNALeu is represented in blue ribbon with the last base Ade76 in sticks; the Ile-AMS analogues bound in the synthetic, and editing sites are shown as salmon and magenta sticks, respectively (carbon atoms).
Ade76 of tRNALeu and Ile-AMS bound in the synthetic site of E. coli LeuRS. The isoleucine part of Ile-AMS shows a gauche conformation. The residues involved in isoleucine discrimination (see text) are represented as sticks.
Same as (B) but with Ile-AMS from the IleRS:Ile-AMS complex (Pdb:1jzq) docked into the LeuRS synthetic site. Isoleucine adopts trans conformation in IleRS, which is not compatible with the LeuRS synthetic site.
Ile-AMS bound in the CP1-editing site of LeuRS, where isoleucine adopts trans conformation. Some of the key residues involved in post-transfer editing of non-cognate amino acids and T252 (involved in leucine rejection) at the editing site of LeuRS are shown in sticks.
Same as (D) but with norvaline post-transfer editing analogue (Nv2AA) docked into the editing site of LeuRS. The position of the aminoacyl part of isoleucine and norvaline is equivalent.