Figure 1. Selection of LOV domains and expression in mammalian cells.
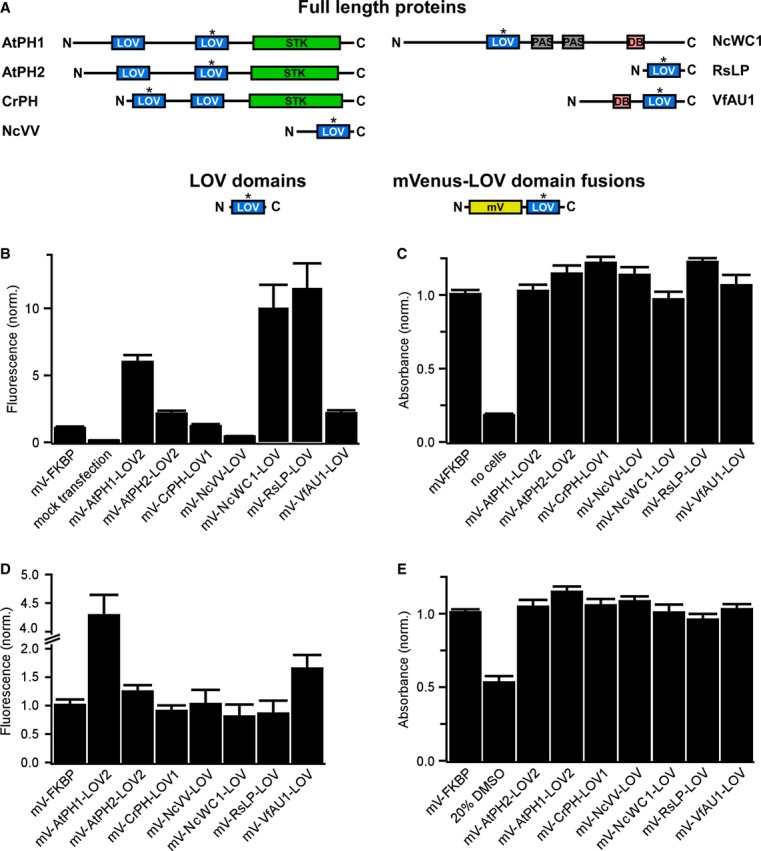
- Domain structure of light-sensing proteins from which LOV domains (highlighted with asterisk) were excised (AtPH1 and AtPH2, Arabidopsis thaliana phototropin 1 and 2; CrPH, Chlamydomonas rheinhardtii phototropin; NcVV, Neurospora crassa vivid; NcWC1, N. crassa white collar 1; RsLP, Rhodobacter sphaeroides ATCC 17025 light-sensing protein; VfAU1, Vaucheria frigida aureochrome1). In these proteins, LOV domains regulate a variety of effector domains (STK, serine/threonine kinase; DB, DNA-binding domain). To test for expression and influence on cell viability in mammalian cells, LOV domains optimized for mammalian codon usage were fused to the fluorescent protein mVenus (mV).
- Fluorescence intensity measurements of human embryonic kidney (HEK) 293 cells transfected with mVenus-LOV domain fusions.
- Viability of HEK293 cells transfected with mVenus-LOV domain fusions.
- Fluorescence intensity measurements of Chinese hamster ovary (CHO) K1 cells transfected with mVenus-LOV domain fusions.
- Viability of CHO K1 cells transfected with mVenus-LOV domain fusions.
Data information: For (B–E): fluorescence and viability were quantified 16–18 h after transfection. Data were normalized to mV fused to the small, robustly folding FK506 binding protein (FKBP). Mean values ± SD for three independent experiments each performed in quadruplicates are shown.
