Table 2.
Tools Used for MeRIP-Seq Differential RNA Methylation Analysis
| Step | Purpose | Tools | Flowchart |
|---|---|---|---|
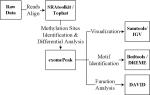
| 1 | Raw data preprocessing and sequence alignment | SRAtoolkit / Tophat [27] |
|
| 2 | RNA methylation sites identification and differential analysis | exomePeak1 [12] | |
| 3 | Motif identification | Bedtools [28] / DREME [29] | |
| 4 | Methylation sites visualization | Samtools [30] / IGV [31] | |
| 5 | Function analysis | DAVID [24] |
The exomePeak package was initially developed as a MATLAB package [12] for RNA methylation site detection from MeRIP-Seq data. It has been recently extended with differential analysis capacity and implemented as an open source R/Bioconductor package.
