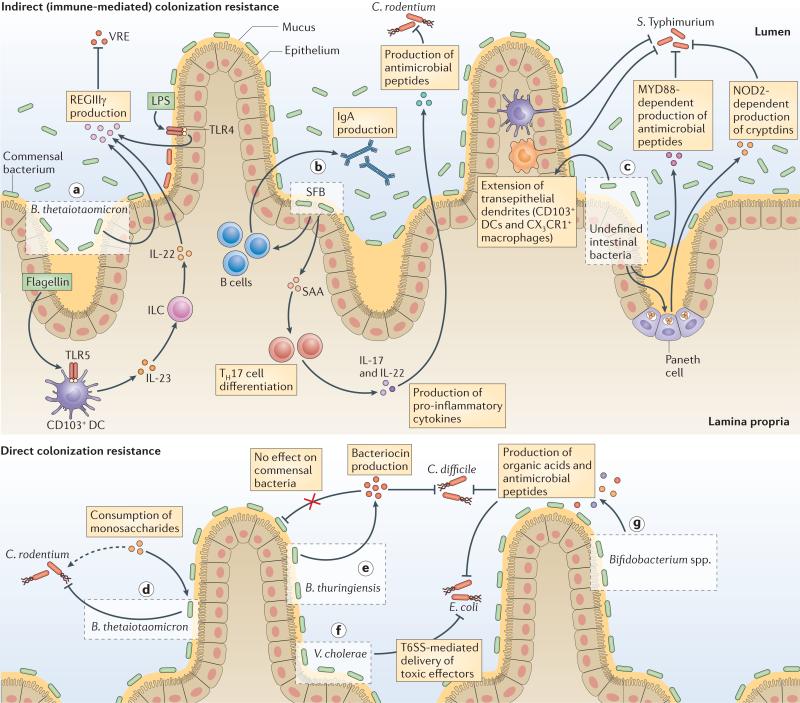
Figure 1. Intestinal bacteria confer indirect (immune-mediated) and direct colonization resistance against enteric pathogens.
The intestinal microbiota enhances colonization resistance to intestinal pathogens by both direct and indirect (immune-mediated) mechanisms of action. Commensal bacterial species and microbial products (green) protect against infection indirectly by activating immune responses that in turn target pathogenic bacteria (red) (parts a–c); for example, Bacteroides thetaiotaomicron enhances expression of the peptidoglycan-binding C-type lectin regenerating islet-derived protein IIIγ (REGIIIγ), which is an antimicrobial peptide that primarily targets and kills Gram-positive bacteria. Microbial products such as lipopolysaccharide (LPS) and flagellin stimulate Toll-like receptor 4 (TLR4)+ stromal cells and TLR5+CD103+ dendritic cells (DCs) to enhance epithelial expression of REGIIIγ, which impairs colonization by Gram-positive vancomycin-resistant Enterococcus spp. (VRE) (part a). Segmented filamentous bacteria (SFB) closely associate with the intestinal epithelium and enhance IgA production by B cells, serum amyloid A (SAA)-dependent T helper 17 (TH17) cell differentiation, pro-inflammatory cytokine production and epithelial production of antimicrobial peptides. These processes confer protection against Citrobacter rodentium (part b). Undefined microbial populations and products activate immune defences, including nucleotide-binding oligomerization domain 2 (NOD2)- dependent cryptdin expression by Paneth cells’ myeloid differentiation primary-response protein 88 (MYD88)-dependent production of antimicrobial peptides and extension of transepithelial dendrites by phagocytic DC populations in the lamina propria. These processes enhance resistance to Salmonella enterica subsp. enterica serovar Typhimurium infection (part c). Other bacteria directly inhibit intestinal pathogens by competing for nutrients or by inducing the production of inhibitory substances (parts d–g); for example, B. thetaiotaomicron consumes carbohydrates used by C. rodentium, which contributes to the competitive exclusion of the pathogen from the intestinal lumen (part d). Bacteroides thuringiensis secretes a bacteriocin that directly targets spore-forming Bacilli and Clostridia, including Clostridium difficile, through an unknown mechanism of action (part e). Gram-negative bacteria, such as Vibrio cholerae, deliver toxic effector proteins directly to Escherichia coli through type VI secretion systems (part f). A variety of Bifidobacterium spp. produce organic acids and peptides that impair growth and adhesion of pathogenic E. coli to enterocytes (part g). Some bacterial populations may inhibit colonization by pathogens through a combination of direct and indirect mechanisms (for example, B. thetaiotaomicron (parts a and d)), or may require other bacteria to carry out antagonistic effects, which complicates the identification and the interpretation of bacterial species that enhance colonization resistance. CX3CR1, CX3C-chemokine receptor 1; IL, interleukin; ILC, innate lymphoid cell.