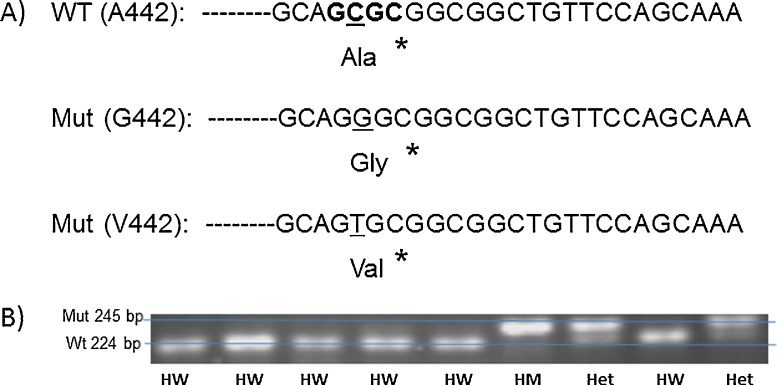
Fig. 4.
(A) Illustration of the principle underlying PCR-RFLP typing of the A442G/V SNP (rs1065761). Primers described by Bierbaum et al. (2006) result in amplification of a 245 bp gene fragment from genomic DNA located in Exon 12 of the CHIT1 gene. The reverse primer introduces a mutation (G → C, denoted by an asterisk) located just two nucleotides downstream of the position of the SNP, introducing an Hinp1L restriction site (G′CGC, bold characters) for the wildtype A442 allele (resulting in a 224 bp and a 21 bp fragment after restriction), but not the A442G/V SNP (uncleaved 245 bp fragment). (B) Example of gel electrophoresis screening for CHIT1 A442G/V SNPs following PCR and digestion with Hinp1L. Higher bands indicate undigested mutant allele A442G or A442V, lower bands indicate A442 wildtype. Single high band indicates homozygous mutant (HM), single lower band indicates homozygous wildtype (HW) and presence of both bands indicates heterozygosity (Het).