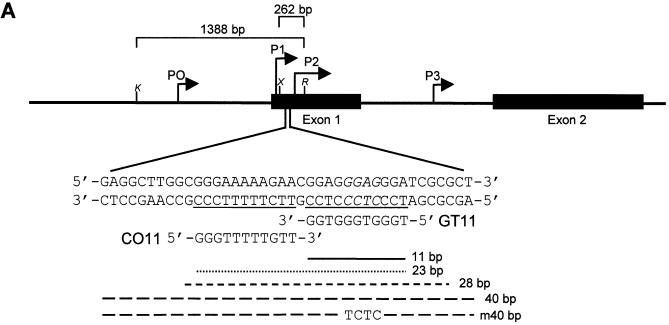
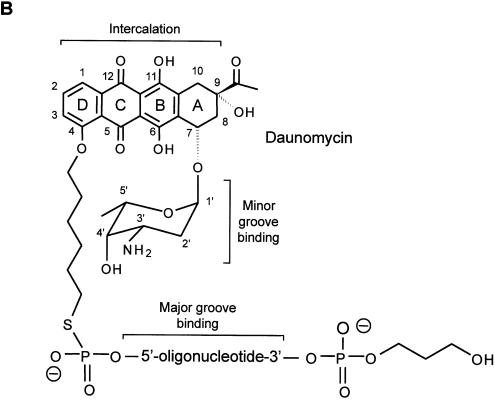
Figure 1.
Map of the c-myc gene and structure of the daunomycin-conjugated TFO. (A) The polypurine:polypyrimidine sequence near the P2 promoter of the c-myc gene and the oligonucleotides GT11 and CO11 are shown. The sites in the target duplex complementary to GT11 and CO11 are underlined. Lines under the sequence indicate the 23, 28 and 40 bp oligonucleotides used in band shift assays and the 11 bp target site. The m40bp is a mutated duplex in which four bases, shown in italics in the target sequence, had been changed to TCTC. Brackets above the gene map indicate the inserts cloned into the pMyc-1388 and pMyc-262, respectively. K, X and R indicate KpnI, XhoI and RsrII restriction sites. (B) Oligonucleotides were modified at the 3′ end with a propanediol tail and linked at the 5′ end to the ring D of the anthraquinone via a hexamethylene bridge. The three domains of the daunomycin-conjugated oligonucleotide are expected to bind by triplex formation in the major groove (oligonucleotide), minor groove binding (amino sugar) and intercalation at the duplex-triplex junction (anthraquinone), respectively.