Figure 4.
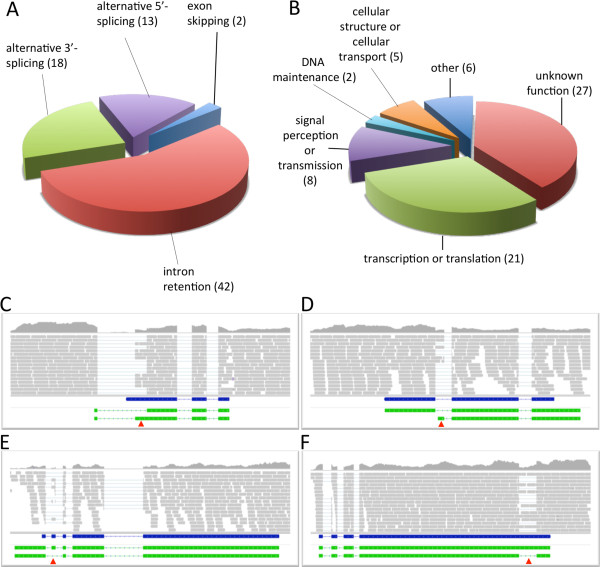
Identification of alternative splicing. A. Overview of types and frequency of putative alternative splicing events in C. graminicola. B. Classification of alternatively spliced genes according to their functions. C. Alternative 3′- or (D) 5′- splice sites were identified by a significant number of reads covering intron sequences at their 3′- or 5′-ends whereas intron retention (E) could be identified by reads covering the entire intron sequence. Exon skipping (F) was identified by a significant number of reads that cover predicted intron sequences, but do not bridge the gap between the alternate exon and adjacent exons. Blue bars visualize the structure of gene models predicted in the current annotation of the C. graminicola genome. Manually improved alternate gene model are illustrated by green bars, grey bars represent individual RNA-Seq reads.
