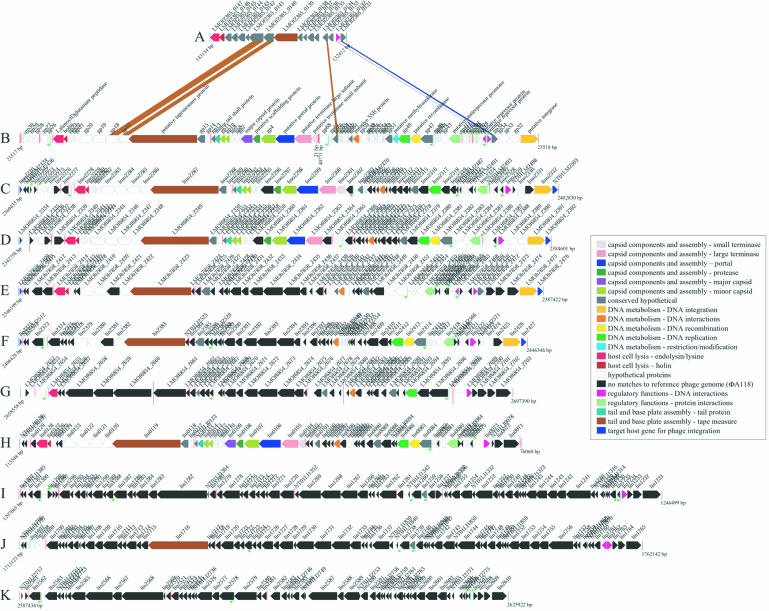
Figure 2.
Putative prophage regions within the genomes of five sequenced Listeria genomes. ORF colors are based on the annotation of the published sequence of L.monocytogenes 1/2a-specific typing phage φA118 (B). All putative prophage ORFs are colored to match φA118 if the protein sequence has a BLASTP P-value cut-off of ≤1 × 10–5, and a percentage identity ≥30% over 75% or more of the length of the query sequence. If there is no match to φA118 based on the above cut-offs, the ORF is colored black. The prophages are denoted as follows: (A) putative monocin region from L.monocytogenes F2365; (B) L.monocytogenes 1/2a-specific typing phage φA118 (Genbank accession NC_003216); (C) putative A118-like prophage φEGDe.2 from L.monocytogenes EGD-e inserted into comK; (D) putative A118-like prophage φF6854.2 from L.monocytogenes F6854 inserted into comK; (E) putative A118-like prophage φH7858.2 from L.monocytogenes H7854 inserted into comK; (F) putative A118-like prophage φ11262.5 from L.innocua CLIP 11262 inserted into comK; (G) putative prophage φF6854.3 from L.monocytogenes F6854 inserted into tRNA-Thr-4; (H) putative A118-like prophage φ11262.1 from L.innocua CLIP 11262 inserted into tRNA-Lys-4; (I) putative prophage φ11262.3 from L.innocua CLIP 11262 inserted into a previously undocumented gene similar to lmo1263; (J) putative prophage φ11262.4 from L.innocua CLIP 11262; and (K) putative PSA-like prophage φ11262.6 from L.innocua CLIP 11262 inserted into tRNA-Arg-4. Putative promoters (green bent arrows) were found in the Listeria putative prophages using the predicted promoter sequences from φA118 (22) and the EMBOSS program fuzznuc with a mismatch of 1. Putative transcriptional terminators (red lollypops) were found using the TIGR program TransTerm (www.tigr.org/software). Contig gaps (sequence or physical) are represented by vertical red lines.