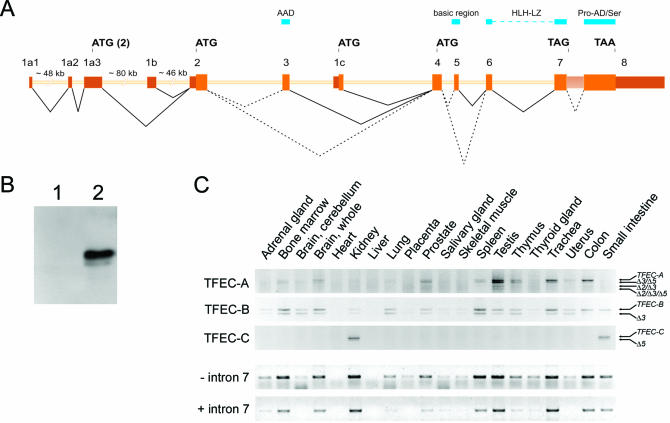
Figure 3.
(A) Schematic representation of the human TFEC gene containing three alternative 5′ exons. For clarity reasons, exon sizes are depicted five times larger than the intron sizes. Relevant start and stop codons are indicated, as well as the regions encoding the AAD, the basic region, the bHLH-LZ and the proline-rich activation domain and serine-rich stretch (Pro-AD/Ser). Dotted lines depict alternative splicing. The accession number of the novel TFEC-C transcript fragment is AJ608795. (B) Western blot analysis of COS-1 cells transfected with empty vector (lane 1) or VSV-tagged TFEC-C cDNA (lane 2). (C) Tissue distribution of the TFEC transcripts determined by RT–PCR analysis. For each of the panels, the various PCR fragments were isolated and sequenced. Next to the full-length TFEC variants, alternatively spliced transcripts were amplified that lacked exon 2 (Δ2), exon 3 (Δ3) and/or exon 5 (Δ5). The lower two panels show RT–PCR products using a forward primer in exon 6, and reverse primers in either exon 8 or intron 7, which depict the presence of TFEC transcripts encoding the complete C-terminus of TFEC or a truncated C-terminus, respectively. In order to enrich for active cytoplasmic transcripts, we used oligo-dT priming in the reverse transcription reaction.