Figure 2.
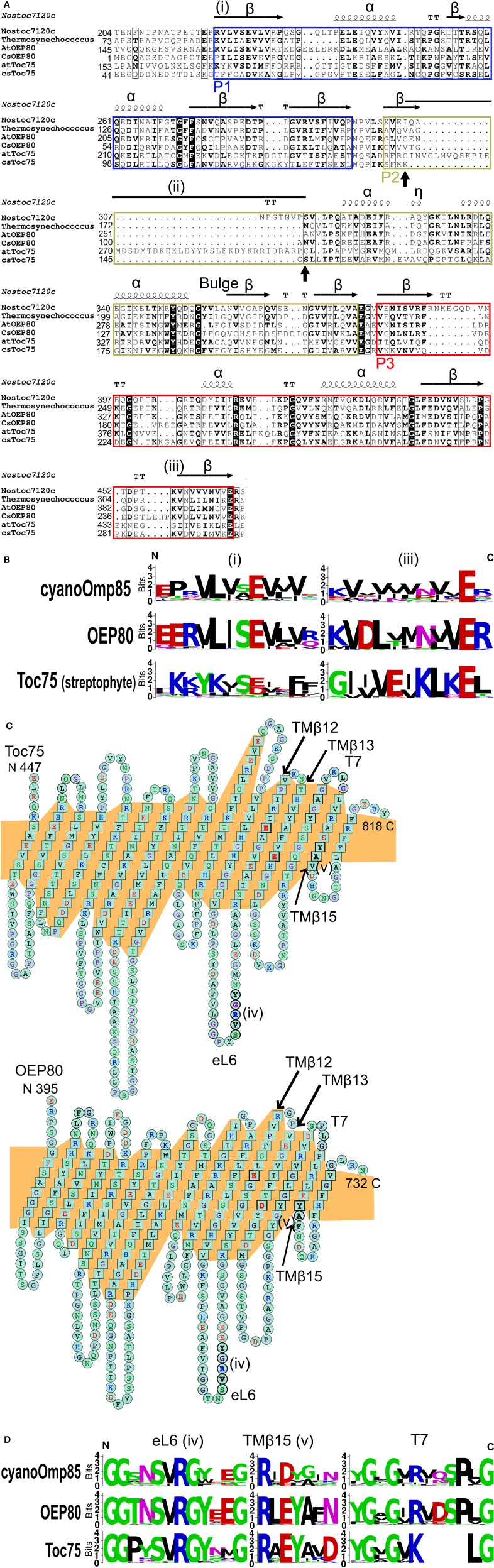
Predicted structures of chloroplast Omp85 homologs. (A) An alignment of the POTRA domains of two cyanoOmp85 orthologs with known structure and four chloroplast Omp85 homologs from the flowering plant A. thaliana (AtOEP80 and atToc75) and the green alga Coccomyxa subellipsoidea (CsOEP80 and csToc75). Shown above the alignment is the resolved secondary structure of Omp85 from Nostoc sp. PCC7120 (Koenig et al., 2010). POTRA domains 1–3 are indicated as P1-3, respectively. The horizontal arrows with β indicate β-strands. The coils show α-helices (indicated with α) or a 3–10 helix (indicated with η) found in the C terminus of the 1st helix of P2. The residues located at turns are indicated with T. Sequences conserved in OEP80 and cyanoOmp85 but diverged in Toc75 (i,iii) as well as an apparent insert at the N terminus of P2 of land plant Toc75 (ii) are indicated. The two vertical arrows below the alignment indicate the positions of Cys residues flanking the insert in Toc75. (B) Weblogos of regions indicated as (i,iii) in panel (A). (C) Predicted transmembrane β-strands of A. thaliana Toc75 and OEP80 generated using the Phyre2 server (Kelley and Sternberg, 2009). The program threaded both proteins onto the known structure of Neisseria gonorrhoeae BamA. The N and C termini of the proteins are indicated at left and right, respectively. Residues in the transmembrane β-strands are indicated in squares and those in turns and loops are in circles. The highly conserved SVRGY motif in eL6 and Y/FA motif in TMβ15 are indicated boldtyped and as (iv,v), respectively. Glu on TMβ12 and Asp on TMβ13 are indicated as boldtyped. The orange background shows the predicted transmembrane region. (D) Weblogos of the conserved sequences shown as (iv,v) in panel (C) and the area surrounding turn 7 (T7) of cyanoOmp85, OEP80, and Toc75 examined in this study.