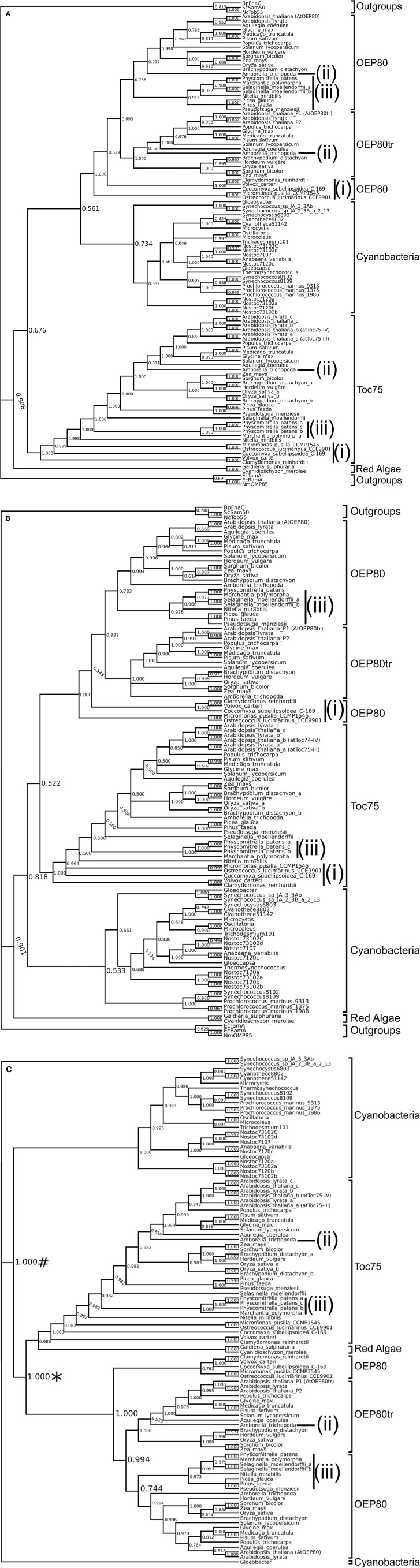
Figure 3.
Phylogenetic trees of Omp85 homologs. Sequences of chloroplast Omp85 homologs (Toc75, OEP80, and OEP80tr), cyanoOmp85, and outgroups from other bacteria and mitochondria were aligned with MAFFT. Phylogenetic inference was done using Bayesian inference. Each tree shown is the 50% majority-rule consensus tree generated for the 3002 trees produced by two runs of two million generations each, sampled every 1000 generations with a 25% burn in. (A) Consensus tree using an alignment of amino acid sequences corresponding to residues 438 to 818 of A. thaliana Toc75 with poorly aligned regions removed by Gblocks. Numbers at nodes represent the proportion of trees in which a clade appeared, interpreted as the posterior probability (PP) of that clade. Discrepancies between our trees and the generally excepted plant species relationships are indicated as lower-case roman numerals (i–iii). (B) Consensus tree using an alignment of amino acid sequences corresponding to residues 648–818 of A. thaliana Toc75 with no interior section removed. (C) Consensus tree using an alignment of amino acid sequences excluding outgroup sequences corresponding to residues 438 to 818 of A. thaliana Toc75. The tree showed the chloroplast Omp85 homologs (*) nesting within cyanobacteria (#).