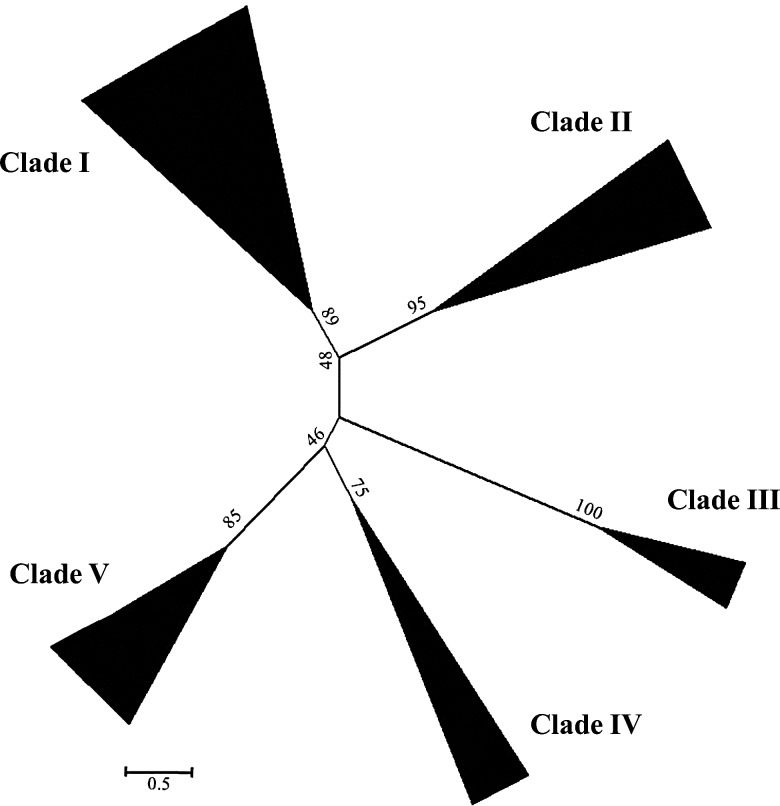
Figure 1.
Phylogenetic relationship among trihelix gene members from the studied species of land plants. The evolutionary history was inferred using the ML method. The bootstrap consensus tree, inferred from 1000 replicates, is taken to represent the evolutionary history of the taxa analysed. Branches corresponding to partitions reproduced in less than 95% bootstrap replicates are collapsed. The evolutionary distances were computed using the number of differences method and are in the units of the number of amino acid differences per sequence.