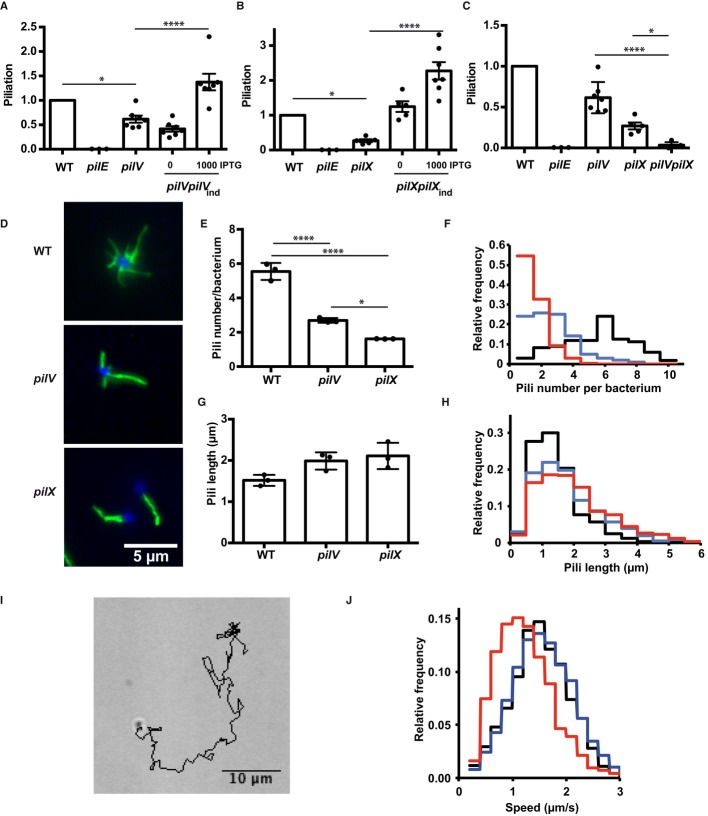
Figure 2. Role of PilV and PilX in pilus biogenesis.
- ELISA analysis of surface-exposed pilin in the pilV mutant and complemented strains. Results are presented relative to the wild-type strain, given the value of 1.
- ELISA analysis of surface-exposed pilin in the pilX mutant.
- ELISA analysis of surface-exposed pilin in the pilVpilX double mutant.
- Immunofluorescence analysis of pili on the surface of the indicated bacterial strains. DAPI is in blue and pilus staining in green (20D9).
- Quantitative determination of the number of pili detected per individual diplococcus. The results of 3 independent experiments each with over 50 bacteria are included.
- Frequency distribution of the number of pili per diplococcus expressed by the wild-type strain (black line), the pilV strain (blue line), and the pilX strain (red line).
- Pili length measurement (μm). The results of 3 independent experiments each with over 50 bacteria are included.
- Frequency distribution of the length of pili expressed by the wild-type, pilV and the pilX strains (same color code as in F).
- A typical track representing bacterial movement resulting from twitching motility over a 2-min period.
- Frequency representation of the different speeds of bacteria for the wild-type, pilV and pilX strains (same color code as in F).
Data information: Data represent mean ± SEM; *P ≤ 0.05; ****P ≤ 0.001.