Figure 3. Loss of NSun2 enhances cellular stress responses.
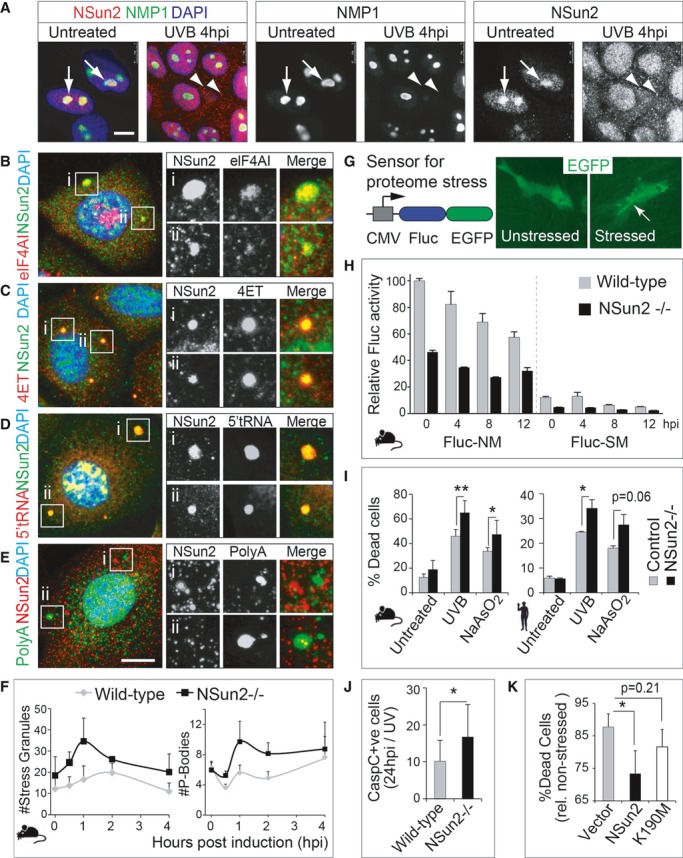
- A Immunofluorescence for NSun2 (red) and NMP1 (green) in untreated (left hand panels) and UV-radiated (right hand panels) human primary keratinocytes. Arrows indicate nucleoli, and arrowheads point to NSun2-positive cytoplasmic granules.
- B–E Co-localisation of NSun2 (green: B–D; red: E) with markers for SGs (eIFAI; red) (B), PBs (4ET; red) (C), 5′ tRNA halves (red) (D) or polyA RNA (green)-containing granules (E) in mouse primary keratinocytes after 4 h of exposure to UV. (i) and (ii) are magnified cytoplasmic granules for the respective stainings.
- F Automated counting of number of SGs (left hand panel) and PBs (right hand panel) per mouse cell after exposure to UV for the indicated hours. Error bars: SEM (n = 3 experiments; 1 experiment: all cells in 30 optical fields).
- G Schematic view of the proteome stress sensor Fluc-GFP construct (left hand panel) and cytoplasmic localisation of GFP-tagged Fluc construct (green) changes to protein aggregates after stress (right hand panel).
- H Luminescence activity of Fluc-NM (not mutated) and Fluc-SM (single mutation) stress reporters in wild-type and NSun2−/− mouse keratinocytes upon UV exposure. Fluc luminescence is normalised to Renilla activity. Error bars, SD (n = 3).
- I Percentage of dead mouse (left hand panel) and human (right hand panel) primary cells in culture expressing (Control) or lacking NSun2 (−/−) 24 h after UV radiation and exposure to NaAsO2.
- J Quantification of cleaved caspase-3-positive cells in back skin of wild-type (WT) and NSun2 knockout (−/−) mice after 24 h of UV radiation.
- K Percentage of dead cells overexpressing an empty vector control (Vector), NSun2 or mutated NSun2 K190M after 24 h of UV treatment.
Data information: In (I–K) error bars represent SD (n ≥ 3). P < 0.05 (*) or P < 0.01 (**). Nuclei are counterstained with DAPI (blue) (A-E). Scale bar: 10 μm (A-E). hpi: hours post-induction. See also Supplementary Fig S4.
Source data are available online for this figure.
