Abstract
Many drugs are effective in the early stage of treatment, but patients develop drug resistance after a certain period of treatment, causing failure of the therapy. An important example is Herceptin, a popular monoclonal antibody drug for breast cancer by specifically targeting human epidermal growth factor receptor 2 (Her2). Here we demonstrate a quantitative binding kinetics analysis of drug-target interactions to investigate the molecular scale origin of drug resistance. Using a surface plasmon resonance imaging, we measured the in situ Herceptin-Her2 binding kinetics in single intact cancer cells for the first time, and observed significantly weakened Herceptin-Her2 interactions in Herceptin-resistant cells, compared to those in Herceptin-sensitive cells. We further showed that the steric hindrance of Mucin-4, a membrane protein, was responsible for the altered drug-receptor binding. This effect of a third molecule on drug-receptor interactions cannot be studied using traditional purified protein methods, demonstrating the importance of the present intact cell-based binding kinetics analysis.
The effectiveness of drugs relies on their specific binding with disease-related receptors1,2. For this reason, determining the drug-receptor binding kinetics has been a major task in drug design and screening3,4. Unfortunately, many drugs are effective only in the early stage of treatment, and patients develop drug resistance over time, causing failure of the therapy. An important example is Herceptin, one of the most successful drugs on the market for treating human epidermal growth factor receptor 2 (Her2) positive breast cancer5,6. The drug inhibits the growth of tumor by binding to overexpressed Her2 in the tumor cell membrane, but most patients develop drug resistance after a short period of treatment5,6. Determining a molecular-scale mechanism of drug resistance is critical to the quest for solutions for drug resistance7. Given the importance of drug-receptor binding in drug actions, it is reasonable to ask: Is the drug resistance caused by the loss or weakening of drug-receptor binding due to steric hindrance8,9 and allosteric modulation10,11,12 by adjacent molecules (alternation hypothesis), or is it due to the development of alternative signaling pathways (bypass hypothesis)13,14,15?
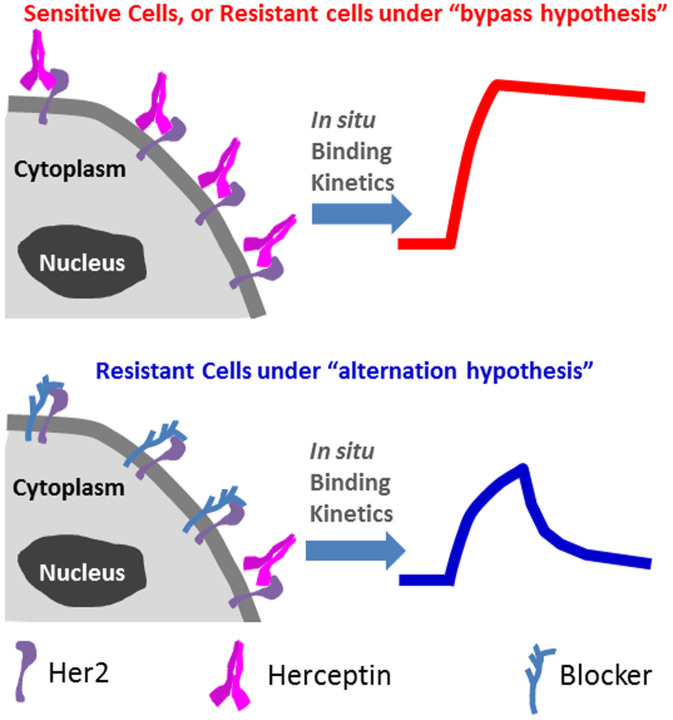
We hypothesized that in situ binding kinetics analysis in intact cells would provide a direct answer to this question, because the “alternation hypothesis” would mean a large change in the binding behaviors of drugs with the receptors in drug-resistant cells while the “bypass hypothesis” would imply no change in the binding kinetics (Fig. 1). Such information cannot be obtained by traditional binding kinetics studies that measure the interactions of drugs with purified membrane receptors immobilized on a solid surface. This is because the purification process isolates the receptors from the native cellular environment where binding modulators are located or bypass pathway developed, and the immobilization may affect the drug-receptor interactions.
Figure 1.
Top: Under “bypass hypothesis”, in situ binding kinetics of Herceptin with resistant cell was not affected. Bottom: “Alternation hypothesis” suggests an altered binding kinetics due to the influences of relevant proteins surrounding Her2.
Here we report a single cell-based quantitative analysis of drug-receptor binding kinetics to investigate the molecular scale origin of drug resistance. We measured the in situ binding kinetics of Herceptin with Her2 in single intact cultured cells and primary cells extracted from breast tumor tissues for the first time with a high resolution surface plasmon resonance (SPR) imaging technique. By doing so, we observed significantly weakened Herceptin-Her2 interactions in Herceptin-resistant resistant cells, compared to those in Herceptin-sensitive cells. Furthermore, we found that the steric hindrance of Mucin-4, a membrane protein, was responsible for the altered drug-receptor binding. This alternation of drug-target interactions by a third molecule in the surrounding environment cannot be observed and studied with the traditional purified protein-based SPR methods, demonstrating the importance of the present intact cell-based binding kinetics analysis.
To date, in situ study of drug-receptor binding in intact cells has been limited to fluorescence labeling-based assays. A popular fluorescence method is kinetic exclusion assay, which measures the concentration of free drugs remaining in the supernatant after the binding equilibrium in cell suspension is achieved16,17,18. This is an end-point assay, and not suitable for extracting the kinetic constants. Furthermore, the use of fluorescent labels is not only labor intensive but may also affect the binding behaviors. Label-free technology, such as SPR, has been developed for studying the binding kinetics of purified proteins in pre-clinical drug screening19,20. We have shown that the interactions of lectin with glycoproteins in single cells can be studied with a SPR imaging setup21. However, the setup lacks the sensitivity and stability to monitor the binding of drugs to low abundance of endogenous receptors.
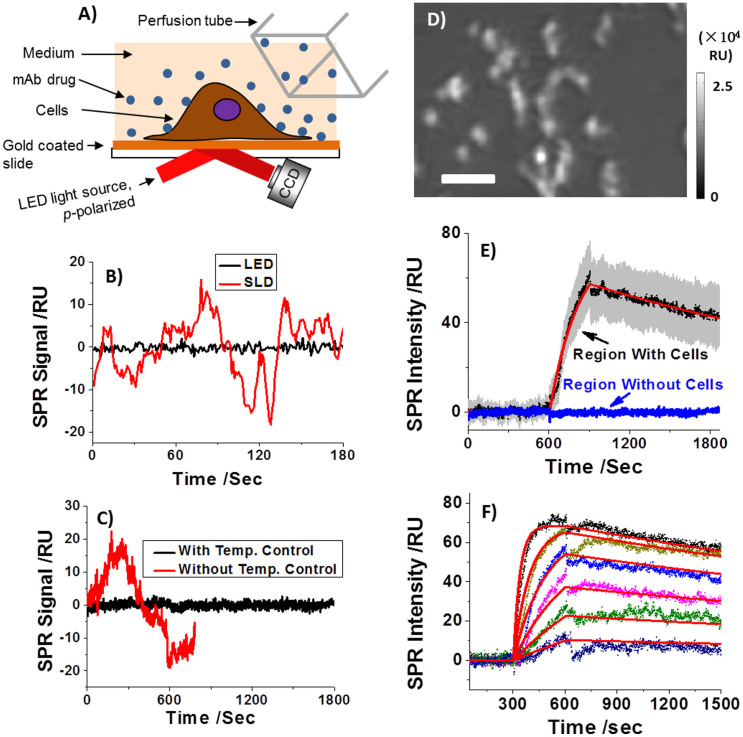
The imaging setup (Fig. 2A) in the present work performs significantly better than the one used in our previous work21, making it possible to study drug binding to endogenous receptors in single cells. The performance improvement was mainly due to optimized light source and implementation of a temperature control system, and mechanical noise reduction stage system. The new setup provides 9 times smaller short-term noise (Fig. 2B), and 11 times smaller long-term drift (Fig. 2C) than those in the previous report21.
Figure 2.
A) Experimental setup. A p-polarized LED light is directed onto a gold-coated glass slide mounted on a SF-11 prim to create SPR on the gold surface, which is captured with a CCD camera. In situ binding kinetics can be extracted from the time lapse SPR image when drug flown over a cell adhered on the gold chip. (B) Short-term noise level when using a superluminescent (SLD, red curves) and a light emitting diode (LED, black curves) as light source. (C) Long-term noise level for a LED light source with and without temperature control. D) Typical SPR image of a few tens of SK-BR3 cells adhered on a gold-coated coverslip. E) The SPR sensorgrams of each individual cells (black: average SPR sensorgram, red: fitted curve, grey background: cell-to-cell variation) and the surrounding regions without cell coverage (blue curve). F) The concentration-dependent SPR sensorgrams and global fitting (red curves). The Herceptin concentration was 21, 8.4, 4.2, 2.1, 1.05 and 0.42 μg/mL from top to bottom curve, respectively. Scale bar, 100 μm.
Fig. 2D shows a typical SPR image of individual SK-BR3 cells attached on gold-coated coverslip. SK-BR3 is a widely used breast cancer cell line and known to express Her2 endogenously in the cell membranes. The overexpression of Her2 in the cell line was further verified by immunofluorescence staining (Supplementary Information). Note that the adherent cells were fixed by 4% paraformaldehyde prior to experiment to minimize the intrinsic micromotions of the cells. To measure the binding kinetics of Herceptin with Her2, PBS buffer was first introduced to flow over the SK-BR3 cells on the surface, and Herceptin dissolved in PBS buffer was then introduced to bind to the cells. After the binding (association) process, PBS buffer was re-introduced to flow over the cells for studying the dissociation process. The entire association and dissociation processes were imaged with the SPR setup, from which sensorgrams of the individual cells were obtained to determine the kinetic constants.
Upon introducing Herceptin, the image intensity in the cell regions increased, indicating the binding of Herceptin to Her2 receptors in the membrane of SK-BR3 cells. The black line in Fig. 2E is an averaged sensorgram over 30 cells, which shows clearly the association (rising) and dissociation (falling) processes of Herceptin on Her2 expressing SK-BR3 cells. The gray background reflects the statistical deviation among all cells. The binding kinetics of Herceptin to SK-BR3 cells can be determined by fitting the average sensorgram based on a 1:1 Langmuir binding model, which is shown by the red curve in Fig. 2E. The corresponding association (kon) and dissociation rate constants (koff) were found to be 1.7 × 105 M−1s−1 and 3.3 × 10−4 s−1, respectively, from which the equilibrium constant (KD) was determined as 1.9 nM. This value is close to the literature report in which Her2-expressing living cells were also used22. No significant intensity change was observed in the regions without the cells (Fig. 2E), indicating the absence of non-specific adsorption of Herceptin to gold regions.
In order to further quantify the variability in the binding kinetics, the Langmuir binding model was used to fit the SPR sensorgram of each individual cell. The histograms of kon, koff and KD for 60 cells are shown in Supplementary Information Section 1. The average kinetic constants are generally consistent with those obtained by fitting the average sensorgram, but the values vary among different cells.
Concentration-dependent kinetic measurement was carried out by sequentially increasing the Herceptin concentration from 0.42 to 21 μg/mL. Note that the cell surface was regenerated for each change of concentration by introducing 100 mM NaOH for 30 seconds (Supplementary Information Section 2). The family of sensorgrams at different Herceptin concentrations, as shown in Fig. 2F, can be fit globally based on the 1:1 Langmuir binding model (red curves), and the corresponding kinetic constants, kon, koff and KD are 1.9 × 105 M−1s−1, 2.3 × 10−4 s−1 and 1.2 nM, respectively. These values are in good agreement with the kinetic constants obtained by fitting each of the individual sensorgrams separately, which further validate the 1:1 Langmuir binding model.
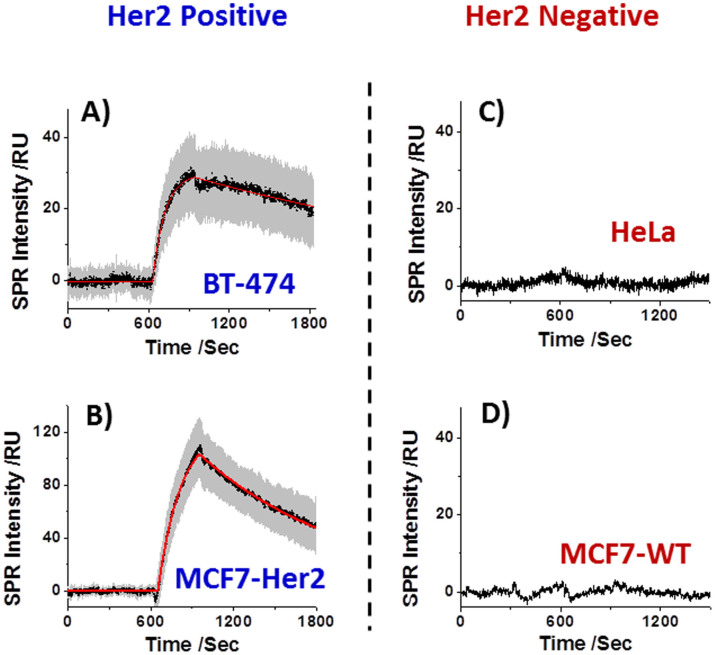
In order to further verify that the SPR binding signal was from the specific interaction between Herceptin drug and the surface-bound Her2 receptors, two Her2-positive (BT474 and MCF7-Her2) and two Her2-negative (MCF7-WT and HeLa) cell lines were included as positive and negative controls, respectively. Like SK-BR3, BT474 is another well-known breast cancer cell line that endogenously overexpresses Her2 in its cell membrane. MCF7-Her2 was developed from Her2-negative wildtype MCF7 cells (MCF7-WT) via gene transduction to stably overexpress Her2, leading to an engineered Her2-positive cell line23. In order to evaluate possible non-specific adsorption of Herceptin onto the cell membrane, Her2-negative MCF7-WT and HeLa, a human epithelial carcinoma cell line, were selected as negative controls. The Her2 expression levels in these cell lines were subsequently validated by the immunofluorescence staining against Her2 (Supplementary Information Section 3).
Herceptin-binding kinetics measurement, similar to those performed for SK-BR3 described above, was performed for each of the cell lines. The average sensorgrams (black curves), and standard deviations (gray background) of BT474 and MCF7-Her2 are shown in Figs. 3A and 3B, respectively. For comparison, the sensorgrams from the negative control cell lines, HeLa (Fig. 3C) and MCF7-WT (Fig. 3D) show flat lines with no sign of Herceptin binding. The sensorgrams for BT474 and MCF7-Her2 can be fit with the 1:1 Langmuir binding model (red curve). The kinetic constants determined from the fitting are listed in Table 1 together with those obtained for SK-BR3 for comparison. An important observation is that the kinetic constants of different cell lines differ by 5 folds, which indicates influences of environments on drug-receptor interactions. Previous studies showed that the binding affinity of Herceptin with intact Her2-containing cell membrane was 50 times weaker than that of Herceptin with free extracellular domain of Her2 in solution22. Molecular interactions occurring at the solution-membrane interface are known to highly depend on membrane properties, such as surface charge, polarity and curvature24. Moreover, cell membrane components that are adjacent to Her2 might also strengthen or weaken its interaction with Herceptin via a positive or negative cooperativity, respectively10,11,12.
Figure 3. SPR sensorgrams when introducing 2.1 μg/mL Herceptin over A) BT474, B) MCF7-Her2, C) HeLa and D) MCF7-WT, respectively.
Table 1. Binding kinetics parameters of different cell lines and clones.
| Cell/Clone | kon 1 × 105 M−1s−1 | koff 1 × 10−4 s−1 | KD nM |
|---|---|---|---|
| SK-BR3 | 1.3 ± 0.4 | 3.4 ± 2.6 | 2.7 ± 2.0 |
| BT474 | 6.2 ± 2.2 | 4.0 ± 2.4 | 0.7 ± 0.4 |
| MCF7-Her2 | 3.6 ± 1.0 | 9.1 ± 4.3 | 2.5 ± 1.6 |
| SK-BR3 (H6)[a] | 2.1 ± 1.0 | 5.4 ± 2.2 | 3.1 ± 2.5 |
| SK-BR3 (C5)[a] | 1.3 ± 0.3 | 5.9 ± 2.6 | 5.5 ± 3.5 |
| SK-BR3 (E8) | 7.4 ± 2.1 | 3.6 ± 1.5 | 0.5 ± 0.2 |
| SK-BR3 (C11) | 3.5 ± 1.7 | 2.4 ± 0.9 | 0.7 ± 0.3 |
[a]Only the effective Her2 population was listed here.
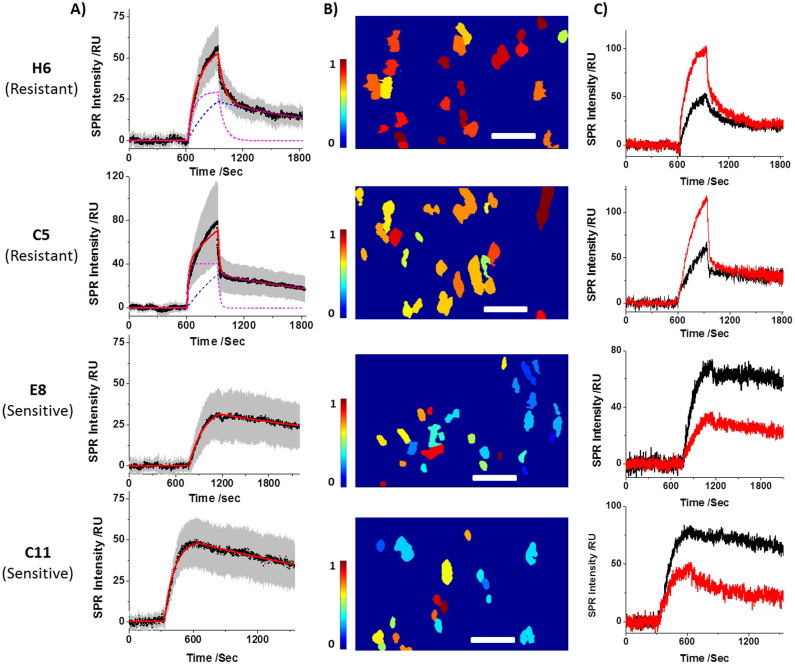
The unique capability for determining drug-receptor binding kinetics in intact cells was applied to study Herceptin resistance mechanism by comparing the Herceptin binding kinetics of Herceptin-sensitive with that of Herceptin-resistant cells. Two Herceptin-sensitive (C11 and E8) and two Herceptin-resistant (H6 and C5) SK-BR3 clones were studied (Supplementary Information Section 5) at a Herceptin concentration of 2.1 μg/mL, which is relevant to the drug concentration in the body used in clinical therapy. The sensorgrams of the four clones are shown in Fig. 4A. The Herceptin-resistant clones, H6 and C5, exhibit two distinct dissociation processes, a fast process followed by a slow process (Figs. 4A, H6 and C5). This behavior is completely different from the dissociation processes in the Herceptin-sensitive clones (Figs. 4A, E8 and C11) and in the other Her2-positive cell lines (Figs. 2D, 3A and 3B), which follow the simple single dissociation kinetics.
Figure 4. Herceptin interactions with SK-BR3 Herceptin-resistant (H6 and C5) and SK-BR3 Herceptin-sensitive (E8 and C11) clones.
A) The average SPR sensorgram of each clone and the corresponding two-constant (H6 and C5) or one-constant (E8 and C11) fitting. B) The resistance index (RI) map of each clone, demonstrating the potency of individual cell being resistant to Herceptin treatment. C) SPR sensorgrams of two individual cells are shown to demonstrate the cell-to-cell variation when interacts with Herceptin. Scale bars, 100 μm.
The two dissociation processes of SK-BR3 H6 and C5 clones can be fit with two exponential functions, each with a different dissociation constant. The kinetics constants of the slow process are listed in Table 1, which are comparable with the dissociation constants of Herceptin-sensitive cells. It indicates that Herceptin remains effective for this population of Her2, which will be thus referred to as the effective population. In contrast, the fast dissociation process is tens of times faster than that found for the regular Herceptin-sensitive cells, indicating the binding of Herceptin to this population of Her2 is greatly weakened in the Herceptin-resistant clones. Consequently, the bound Herceptin has much shorter residence time and can be quickly washed off. This population is expected to be ineffective for Herceptin treatment, and will be referred to as ineffective population.
The sensorgrams from the individual cells exhibit a large variability in the binding kinetics as shown by the gray area in Fig. 4A, indicating that the degree of Herceptin-resistant is different for different cells. This heterogeneity can be quantified with a resistance index (RI) based on the drug binding kinetics, which is the ratio of the weakly bound Herceptin (the ineffective population of Her2) to all the bound Herceptin, including both weakly and strongly bound drug molecules (both ineffective and effective populations of Her2). When RI = 1, all the drug molecules are weakly bound to the cell, and the cell is most drug resistant. On the other hand, when RI = 0, all the drug molecules are strongly bound to the cell, and the cell is least drug resistant (Supplementary Information Section 6).
RI for each of the individual cells was mapped from the SPR intensities at the end of association and 10 minutes after dissociation. The former is proportional to the total bound Herceptin on the cell, and latter reflects the population of the strongly bound Herceptin. Figs. 4B show drug resistant maps of individual cells for H6, C5, E8 and C11, where each spot reflects the morphology of a single cell and the color of each cell represents the RI value. From the maps, the average RI values of H6, C5, E8 and C11 were determined to be 0.78 ± 0.11, 0.58 ± 0.09, 0.31 ± 0.18, and 0.29 ± 0.13, respectively, from which the degree of Herceptin resistance of these clones is in the order of H6 > C5 > E8 ≈ C11. From the colors of the individual cells for each clone in Fig. 4B, the large cell-to-cell variability in the Herceptin-binding within the same clone can be easily visualized. More detailed analysis of the variability in the binding kinetics is shown in Fig. 4C, which plot the sensorgrams of two exemplary cells for each of the clones.
It has been pointed out that the Herceptin binding sites of Her2 in the Herceptin-resistant cells were probably blocked by certain membrane proteins, such as Mucin-4 (MUC4)8. However, a specialized cell line was used in that research and no direct evidence on the altered binding kinetics was provided. Further more, the end-point fluorescence assay could not identify the ineffective population because the drug can be easily washed off. The kinetics study here suggests the co-existence of two populations, and that the binding sites are still available for Herceptin, however, the bound Herceptin to these binding sites dissociates quickly.
In order to examine the role of MUC4 in the ineffective population, the following experiments were carried out. 1) The expression levels of MUC4 of H6, C5, E8 and C11 were determined by immunofluorescence staining (Fig. 5A). The results show that the MUC4 expression levels in the Herceptin-resistant clones are indeed much higher than those in the Herceptin-sensitive clones (Supplementary Information Section 7). 2) The abundance levels of Her2 in all the clones were studied with immunofluorescence staining by using another anti-Her2 polyclonal antibody that recognizes different binding sites in Her2. The results confirm the finding from the SPR kinetic data that overall Her2 expression level does not significantly decrease in Herceptin-resistant clones (Supplementary Information Section 8). 3) Dual-staining of Her2 (using Herceptin as primary antibody) and MUC4 for a given Herceptin-resistant C5 clone was carried out (Fig. 5B and Supplementary Information Section 9), showing that the regions with more MUC4 tend to have less Herceptin bound to Her2, and vice versa. Such negative co-localization was observed in lots of the studied cells with varied extents, this particular cell shown in Fig. 5B was selected for best presentation. Note that the Her2 staining result in Fig. 5B (green) only represents the distribution of effective Her2 population rather than the total Her2 population, since those regions with ineffective Her2 population would lose all the Herceptin during the rinsing stage in the assay. The present study suggests that MUC4 interacts with some of the Her2 in the drug resistant cells, which weakens the binding of Herceptin to Her2. This is believed to be associated with the ineffective Her2 population observed in the SPR sensorgrams. The experimental data in the present work are consistent with the alternation hypothesis. Additional studies are needed to further elucidate the mechanism.
Figure 5. Molecular-scale mechanism of ineffective Her2 population.
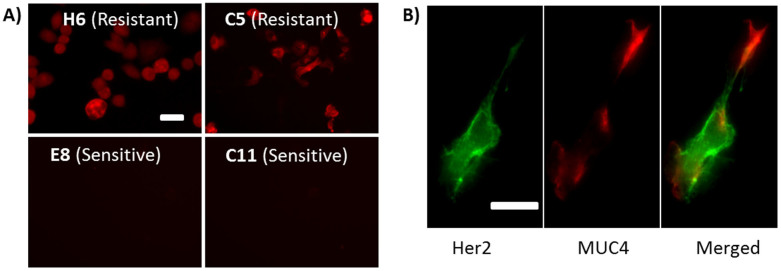
A) Resistant clones (H6 and C5) overexpress MUC4 while sensitive clones (E8 and C11) do not. B) Dual-staining of Her2 and MUC4 in one single cell showing negative co-localization for Her2 and MUC4. Similar features were observed in multiple cells, but only a representative one was shown here. Scale bars, 50 μm.
In conclusion, a single cell-based kinetic analysis approach was developed to elucidate the molecular scale origin of drug resistance. The approach uses a label-free SPR imaging technique to image and quantify the interactions of drugs with low abundance endogenous receptors in intact cells. Using the approach, the binding kinetics of Herceptin, an anti-tumor drug, with its receptor, Her2, in both drug-sensitive and drug-resistant cells were quantified. The study revealed two distinct populations of Her2 in the Herceptin-resistant cells. The first one has binding kinetics similar to that of Herceptin-sensitive cells, but the second population has a dissociation process tens of times faster, indicating a short residence time of bound Herceptin. The presence of the second population is likely responsible for the drug-resistance. This important information is not available from any other studies carried out to date. The method and technique developed here are expected to be applicable to study other drug-receptor interactions for screening of drugs, and for elucidation of drug resistance mechanism.
Methods
Materials
Herceptin was obtained from Genentech Inc. (San Francisco, CA) as powder. The Herceptin stock solution was prepared by dissolving 42 mg powder in 1 mL pure water, reaching an effective mAb concentration of 21 mg/mL by following the manufacturer's instruction. The Herceptin solution used in the present work was prepared by correspondingly diluting the stock solution with PBS. 4% Paraformaldehyde was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Alexa Fluor 488 goat anti-human IgG1 (γ1), Alexa Fluor 555 goat anti-mouse IgG1 (γ1) and MUC4 mouse mAb (clone 1G8) were purchased from Invitrogen Inc. (Carlsbad, California). All the reagents were analytical grade and purchased from Sigma-Aldrich, except where otherwise stated. Deionized water was used to prepare all the buffers.
SK-BR3 Herceptin sensitive and resistant clones selection
SK-BR3 Herceptin sensitive and resistant clones were developed by Dr. Laura Gonzalez in LaBaer's lab. In brief, SK-BR3 cells were diluted in order to have at least one cell/well and plated into 10 plates of 96 multiwells and grown for approximately one month. When cells reach confluence, they were trypsinized and condensed into 2 plates. After condensing all the subclones that grew, they were grown again to confluence and split into 3 different plates: one as archive, one as a control and the last one treated with Herceptin. After 6 days post-treatment, cell proliferation was measured by resazurin. When survival of cells was inhibited by at least 50% compared to non-treated cells, we considered them as Herceptin-sensitive, whereas if cells continue to grow in presence of Herceptin with numbers at least 80% of those for non-treated cells, we consider them as Herceptin-resistant (Fig. S5).
MCF7-Her2 transfection
Retroviral particles were prepared to transduce MCF7 cells to stably express Her2, as described before23. In brief, packaging cell line HEK 293T was transfected with high quality plasmid DNA of retroviral construct (pJP1520-Her2), mixed with two other plasmids (retroviral gag-pol and VSV-G). Virus was then harvested daily for 4 days, aliquoted and storage at -80°C. Then MCF7 cells were seeded in 6 well format plates and transduced using these retroviruses which has puromycin acetyl transferase gene to allow the selection of infected cells. Cells were kept in the presence of puromycin (1 μg/ml) until reach confluence and expanded in bigger flasks until stable cell line was established.
Cell line culture
SK-BR3, BT-474, MCF7-WT cell lines were kindly provided by Dr. LaBaer and HeLa cell line was obtained from the American Type Culture Collection (Rockville, MD). All the cells were cultured in a humidified incubator at 37°C with 5% CO2 and 70% relative humidity. SK-BR3 parental and four clones were grown in RPMI-1640 (Invitrogen, Carlsbad, California) with 10% FBS (Invitrogen, Carlsbad, California), penicillin and streptomycin (BioWhittaker, Basel, Switzerland). The resistance clones were cultured in the presence of additional 200 μg/mL Herceptin. BT-474, MCF7-WT, MCF7-Her2 and HeLa cells were grown in DMEM (Invitrogen, Carlsbad, California) with 10% FBS, penicillin and streptomycin. MCF7-Her2 cells were cultured in the presence of 1 μg/mL puromycin. Cells were passaged with 0.05% trypsin and 0.02% ethylenediaminetetraacetic acid in a Hank's balanced salt solution (Sigma-Aldrich, St Louis, Missouri) when they were approximately 75% confluent. The cells were fixed prior to the kinetic measurement in order to minimize noises caused by the micromotions of live cells. The fixation to cells was performed by incubating with a 4% paraformaldehyde solution for 15 minutes at room temperature.
SPRi setup
The measurement was carried out with homemade SPR imaging setup, in which p-polarized light from a 670 nm light-emitting diode (L7868-01, Hamamatsu, Japan) was directed through a triangle SF-11 prism onto the gold-coated BK-7 glass (chip) placed on the prism. The LED light source was mounted on a temperature-controlled mounting socket (LDM21, Thorlabs, Newton, USA). The reflected beam produced an SPR image, which was captured by a CCD camera (Pike F032B, Allied Vision Technology, Newburyport, USA). The optics included 10× magnification, allowing for imaging of multiple individual cells. A Flexi-Perm silicon sample cell (Greiner Bio-One) was placed on top of the gold chip to for cell culture. To promote cell adhesion, each gold chip was modified with a collagen layer by incubating the chip in 50 μL of 10 μg/mL collagen solution for 2 hours. The chip was then rinsed with deionized water twice prior to cell seeding using 5,000 cells in 250 μL of growth medium. To allow the cells to attach and grow on the chip surface, the chip was incubated in the growth medium for 3 days. The growth medium was then replaced with PBS buffer solution, and the cells were fixed by 4% paraformaldehyde solution before each for binding kinetics experiment. All the SPR measurements were performed at room temperature (~25°C).
Immunofluorescence
The immunofluorescence images were obtained at an inverted fluorescence microscope at 60× (Fig. 5B only) or 10× (otherwise) magnification (Olympus IX81). A 150 W mercury lamp was used as the light source. For Her2 staining, the fixed cells were incubated with 10 μg/mL Herceptin solution for 30 minutes, followed by another 30 minutes incubation with Alexa Fluor 488 goat anti-human IgG1 (γ1) after 3 times rinses with PBS at room temperature. A commercial anti-Her2 polyclonal antibody from rabbit (#2242, Cell Signaling Technology) and corresponding Alexa Fluor 488 goat anti-rabbit IgG (A-11008, Life Technologies) were used in Fig. S6. The fluorescence image was captured after the well was rinsed twice by PBS buffer. A set of optical filters (Ex420-480/Em515) was applied for Her2 staining. For MUC4 immunofluorescence, the fixed cells were incubated with MUC4 mouse mAb (clone 1G8) for 30 minutes, then rinsed 3 times in PBS, followed by the incubation with Alexa Fluor 555 goat anti-mouse IgG1 (γ1) for another 30 minutes at room temperature. The fluorescence image was captured after the well was rinsed twice by PBS buffer. A set of optical filters (Ex535/Em565) was applied for MUC4 immunofluorescence.
Flow system
A gravity-based multichannel drug perfusion system (SF-77B, Warner Instruments, Connecticut) was used to control the local solution that surrounded the target cell. The typical transition time between different solutions was about 1–2 seconds.
Author Contributions
W.W., Y.L., L.G. and S.E. designed and performed the experiments. S.W. contributed the SPRi instrumentation. W.W., L.G., X.Y., S.Z., H.Y.C., J.L. and N.J.T. discussed the results. H.Y.C. helped with the experimental materials. W.W., L.G. and N.J.T. wrote the paper. N.J.T. conceived the experiment and supervised the project.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Genentech Inc. for providing the Herceptin used in the present research, and the financial supports from National Natural Science Foundation of China (NSFC, Grants No. 21327008, 21405080), Natural Science Foundation of Jiangsu Province (BK20140592), National Institute of Health (NIH, 8R21GM103396-03 and #1R01GM107165-01A1), National Science Foundation (BIO-1151005) and the Breast Cancer Research Foundation.
References
- Ehrlich P. Address in pathology on chemotherapeutics: Scientific principles, methods, and results. Lancet 2, 445–451 (1913). [Google Scholar]
- Adams G. P. & Weiner L. M. Monoclonal antibody therapy of cancer. Nature Biotechnol. 23, 1147–1157 (2005). [DOI] [PubMed] [Google Scholar]
- Copeland R. A., Pompliano D. L. & Meek T. D. Opinion - Drug-target residence time and its implications for lead optimization. Nature Rev. Drug Disc. 5, 730–739 (2006). [DOI] [PubMed] [Google Scholar]
- Swinney D. C. The role of binding kinetics in therapeutically useful drug action. Curr. Opin. Drug Disc. Develop. 12, 31–39 (2009). [PubMed] [Google Scholar]
- Nahta R., Yu D. H., Hung M. C., Hortobagyi G. N. & Esteva F. J. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nature Clin. Pract. Oncol. 3, 269–280 (2006). [DOI] [PubMed] [Google Scholar]
- Valabrega G., Montemurro F. & Aglietta M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Annal. Oncol. 18, 977–984 (2007). [DOI] [PubMed] [Google Scholar]
- Montagut C. et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature Med. 18, 221–223 (2012). [DOI] [PubMed] [Google Scholar]
- Nagy P. et al. Decreased accessibility and lack of activation of ErbB2 in JIMT-1, a herceptin-resistant, MUC4-Expressing breast cancer cell line. Cancer Res. 65, 473–482 (2005). [PubMed] [Google Scholar]
- Price-Schiavi S. A. et al. Rat MUC4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for Herceptin resistance. Int. J. Cancer 99, 783–791 (2002). [DOI] [PubMed] [Google Scholar]
- Christopoulos A. & Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54, 323–374 (2002). [DOI] [PubMed] [Google Scholar]
- Christopoulos A. Allosteric binding sites on cell-surface receptors: Novel targets for drug discovery. Nat. Rev. Drug Disc. 1, 198–210 (2002). [Google Scholar]
- Kenakin T. & Miller L. J. Seven Transmembrane Receptors as Shapeshifting Proteins: The Impact of Allosteric Modulation and Functional Selectivity on New Drug Discovery. Pharmacol. Rev. 62, 265–304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kute T. et al. Development of Herceptin resistance in breast cancer cells. Cyto. Part A 57A, 86–93 (2004). [DOI] [PubMed] [Google Scholar]
- Lu Y. H., Zi X. L., Zhao Y. H., Mascarenhas D. & Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 93, 1852–1857 (2001). [DOI] [PubMed] [Google Scholar]
- Nahta R., Takahashi T., Ueno N. T., Hung M. C. & Esteva F. J. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res. 64, 3981–3986 (2004). [DOI] [PubMed] [Google Scholar]
- Darling R. J. & Brault P. A. Kinetic exclusion assay technology: Characterization of molecular interactions. Assay Drug Develop. Technol. 2, 647–657 (2004). [DOI] [PubMed] [Google Scholar]
- Xie L. et al. Measurement of the functional affinity constant of a monoclonal antibody for cell surface receptors using kinetic exclusion fluorescence immunoassay. J. Immuno. Methods 304, 1–14 (2005). [DOI] [PubMed] [Google Scholar]
- Rathanaswami P., Babcook J. & Gallo M. High-affinity binding measurements of antibodies to cell-surface-expressed antigens. Anal. Biochem. 373, 52–60 (2008). [DOI] [PubMed] [Google Scholar]
- Ernst R. E., High K. N., Glass T. R. & Zhao Q. Determination of Equilibrium Dissociation Constants. in Therapeutic Monoclonal Antibodies: From Bench to Clinic. (ed. An, Z.) (John Wiley & Sons, Inc., Hoboken, NJ, USA; 2009). [Google Scholar]
- Neumann T., Junker H. D., Schmidt K. & Sekul R. SPR-based fragment screening: Advantages and applications. Curr. Top. Med. Chem. 7, 1630–1642 (2007). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nature Chem. 4, 846–853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troise F., Cafaro V., Giancola C., D'Alessio G. & De Lorenzo C. Differential binding of human immunoagents and Herceptin to the ErbB2 receptor. FEBS J. 275, 4967–4979 (2008). [DOI] [PubMed] [Google Scholar]
- Pearlberg J. et al. Screens using RNAi and cDNA expression as surrogates for genetics in mammalian tissue culture cells. Cold Spring Harb. Symp. Quant. Biol. 70, 449–459 (2005). [DOI] [PubMed] [Google Scholar]
- Knezevic J. et al. Quantitation of Affinity, Avidity, and Binding Kinetics of Protein Analytes with a Dynamically Switchable Biosurface. J. Am. Chem. Soc. 134, 15225–15228 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information