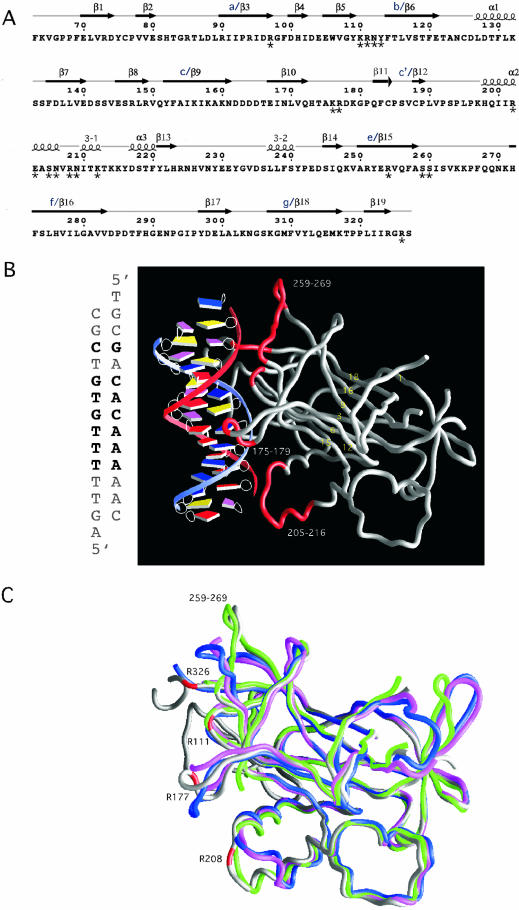
Figure 1.
(A) The sequence of the DNA-binding domain of Ndt80 (1M6U) (11) with the indicated secondary structures that were derived from ESPript (26). Secondary structural elements are labeled as previously reported. For the sake of comparison, alternate names (a–g) for structural elements in the β-sandwich core have also been included (10). Residues interacting with the DNA that have been mutated are indicated with an asterisk below the amino acid. Additional residues that we have mutated (see Table 1) that are not included in this schematic include K50, K54, I55, P57 and R58. (B) The crystal structure of the DNA-binding domain of Ndt80 bound to MSE DNA. The sequence of the MSE DNA that is bound to Ndt80 in the crystal structure is shown on the left. The protein molecule is shown as a worm rendering and the DNA as a spline with boxes representing individual bases (25). The base pairs that are in common with the SMK1-MSE DNA used in our assays are in bold letters. The β-strands that are in the β-sandwich core are labeled in yellow. Residues in loop regions that interact directly with the MSE DNA are highlighted in red. (C) A comparison of the Ndt80 peptide backbone structures shown as worm renderings in the absence (PDB: 1M6U, blue; 1M7U, pink; 1MN4 green) and presence of DNA (PDB: 1MNN, gray). Representative residues that contact the DNA in the crystal structure of Ndt80 in complex with DNA are highlighted in red. Regions of the protein that differ significantly in the structures compared include residues 175–180, 205–216, 259–269 and the C-terminus of the protein including residue 326.