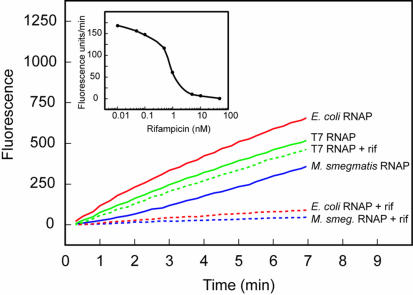
Figure 5.
Use of molecular beacons to assess the inhibition of different RNA polymerases by rifampicin. Each transcription reaction contained either 1 U/µl bacteriophage T7 RNA polymerase, 0.05 U/µl E.coli RNA polymerase or 1 µl M.smegmatis RNA polymerase (4). Each reaction contained DNA templates possessing a promoter specific for the RNA polymerase (RNAP) that was present and molecular beacons that were specific for the resulting transcripts. The kinetic course of reactions carried out in the absence of rifampicin (continuous lines) was compared to the kinetic course of otherwise identical reactions carried out in the presence of 5 nM rifampicin (rif) (dotted lines). The results shown in the inset illustrate the relationship between the initial rate of transcription catalyzed by E.coli RNA polymerase and the logarithm of rifampicin concentration. T7-sinR-poly(A) was used as template in the T7 RNA polymerase reactions and sinP3-sinR-poly(A) was used as template in both the E.coli RNA polymerase reactions and the M.smegmatis RNA polymerase reactions. All reactions were monitored using molecular beacon MB-poly(A)-FAM-2′-O-methyl.