Figure 4.
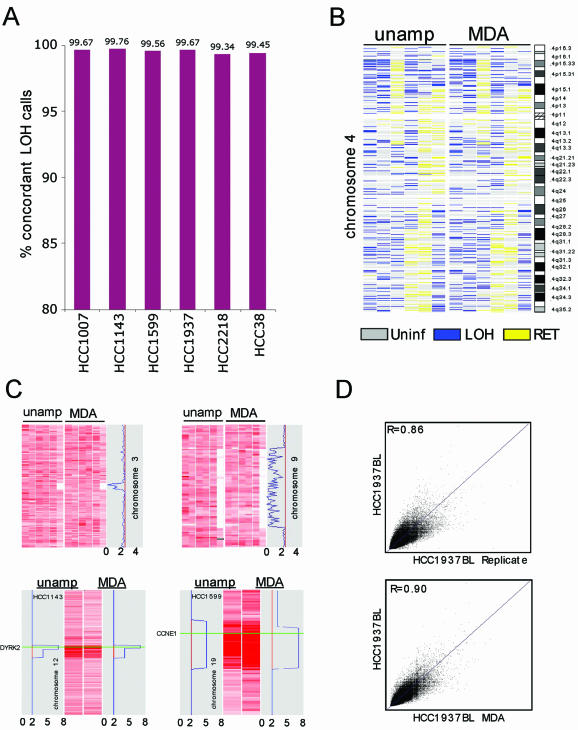
Detection of LOH, homozygous deletions and gene amplification after φ29MDA. (A) Concordance of LOH detection in cancer/normal samples before and after φ29MDA. HCC1007 and HCC1143 tumor/normal pairs and HCC1599 BL were denatured prior to amplification. (B) LOH map of chromosome 4 representative of whole genome LOH maps from six breast cancer/normal pairs. Informative SNP loci showing retention of heterozygosity are shown in yellow, uninformative SNPs (i.e. homozygous or no-call in normal) in gray and SNP alleles undergoing LOH in blue. The first two columns correspond to denatured tumor/normal DNA samples. For the third column, only the normal DNA was denatured. The remaining three columns correspond to all non-denatured tumor/normal pairs. (C) Detection of homozygous deletions and gene amplification. SNP signal intensity is displayed as rows where intensity is normalized on a scale of 0 (white) to 6 (dark red). The upper panels show two representative regions of homozygous deletion on chromosomes 3 and 9 in six breast cancer/normal pairs. The first two columns correspond to tumor samples denatured prior amplification. The lower panel shows representative regions of amplification on chromosomes 12 and 19. Only the individual cancer cell line samples showing gain are shown. Copy number loss or gain is indicated by the blue line in the gray box. HCC1143 was denatured prior amplification whereas HCC1599 was not. (D) Representative SNP hybridization intensity concordance plots. Hybridization intensities from unamplified DNA are plotted against corresponding intensities from a second replicate hybridization of unamplified DNA (top) and from whole genome amplified DNA (bottom). The best fit linear correlation is represented in blue. Correlation coefficients (top left corners), representing signal intensity concordance rates, are similar in both cases. These data are representative examples of all pair-wise MDA/unamplified and unamplified/replicate comparisons examined in this manner. The mean R values for these comparisons are presented in the text.