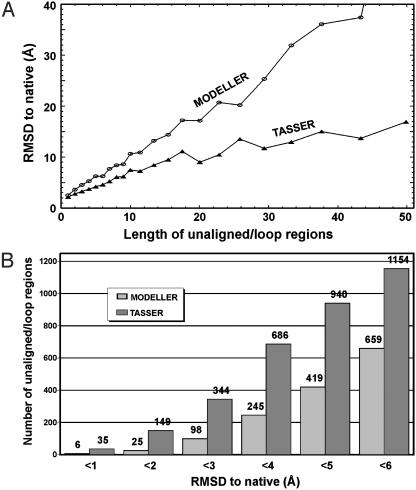
Fig. 5.
(A) Average rmsd to native of all unaligned/loop regions by tasser and modeller (3) as a function of loop length. The rmsd is calculated based on the superposition of up to five neighboring stem residues on both sides of the loop. (B) Histogram of the rmsd for the unaligned/loop regions with ≥4 residues (1,968 in total) modeled by tasser and modeller.