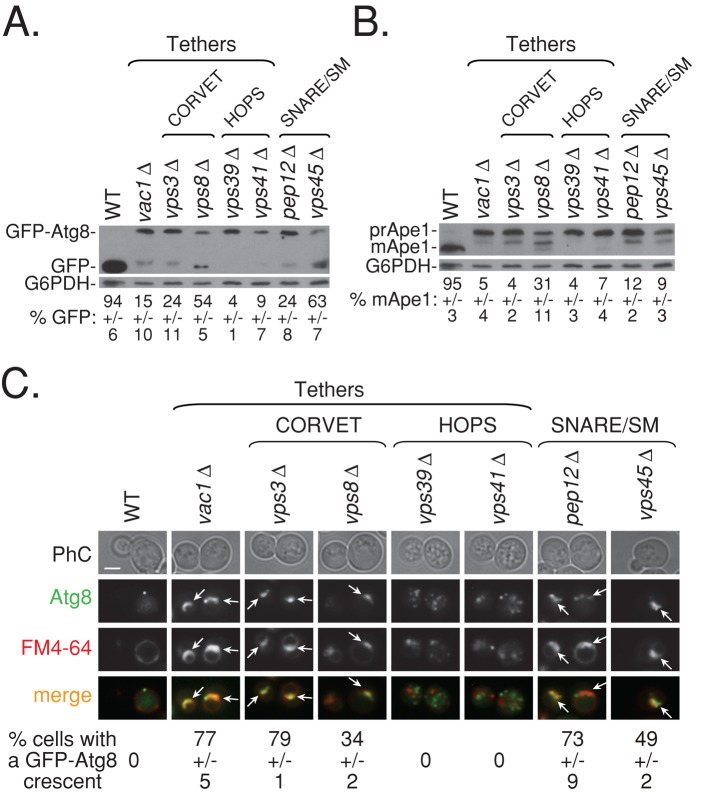
FIGURE 7:
Cells deleted for factors that function downstream of Vps21 exhibit autophagic phenotypes similar to or greater than those of vps21∆ mutant cells. Cells were deleted for CORVET-specific components, Vps3 and Vps8; HOPS-specific components, Vps39 and Vps41; and other class D Vps members, Pep12, Vac1, and Vps45. Wild-type and mutant cells were shifted to SD-N medium and their autophagic phenotypes, GFP-Atg8 processing (A), Ape1 processing (B), and GFP-Atg8 accumulation (C), were determined as described in the legends to Figures 1 and 2. All mutant cells are defective in GFP-Atg8 (A) and Ape1 (B) processing (same blot was probed with the antibodies sequentially), with vps8∆ and vps45∆ mutant cells showing the weakest phenotypes (p < 0.04 when compared with wild type). (C) Mutant cells deleted for Vps21-related factors, Vps3, Vps8, Vac1, and Vps45, and the SNARE Pep12, but not Ypt7-related factors, Vps39 and Vps41, accumulate GFP-Atg8 crescents (>70% for vps3∆, pep12∆, and vac1∆, and ∼35% for vps8∆ and vps45∆; number of cells visualized for each strain, >700 cells). ± represents STD; bar, 2 μm; arrows point to GFP-Atg8 crescents. Results represent three independent experiments.