Abstract
Purpose
The presence of diffuse anaplasia in Wilms tumours (DAWT) is associated with TP53 mutations and poor outcome. As patients receive intensified treatment, we sought to identify whether TP53 mutational status confers additional prognostic information.
Patients and Methods
We studied 40 patients with DAWT with anaplasia in the tissue from which DNA was extracted and analysed for TP53 mutations and 17p loss. The majority of cases were profiled by copy number (n = 32) and gene expression (n = 36) arrays. TP53 mutational status was correlated with patient event-free and overall survival, genomic copy number instability and gene expression profiling.
Results
From the 40 cases, 22 (55%) had TP53 mutations (2 detected only after deep-sequencing), 20 of which also had 17p loss (91%); 18 (45%) cases had no detectable mutation but three had 17p loss. Tumours with TP53 mutations and/or 17p loss (n = 25) had an increased risk of recurrence as a first event (p = 0.03, hazard ratio (HR), 3.89; 95% confidence interval (CI), 1.26–16.0) and death (p = 0.04, HR, 4.95; 95% CI, 1.36–31.7) compared to tumours lacking TP53 abnormalities. DAWT carrying TP53 mutations showed increased copy number alterations compared to those with wild-type, suggesting a more unstable genome (p = 0.03). These tumours showed deregulation of genes associated with cell cycle and DNA repair biological processes.
Conclusion
This study provides evidence that TP53 mutational analysis improves risk stratification in DAWT. This requires validation in an independent cohort before clinical use as a biomarker.
Introduction
Wilms tumour (WT) or nephroblastoma, the most frequent renal tumour of childhood, affects around 1 in 10,000 children before the age of 15 years. Most patients with WT in the Western world are treated within prospective clinical trials conducted by either the International Society of Paediatric Oncology - Renal Tumor Study Group (SIOP-RTSG, Europe) or the Children's Oncology Group (COG, North America) that each report long-term survival rates of over 85% [1]–[8]. However, a substantial minority (∼25%) respond poorly or relapse with current therapies and approximately 50% of these children will succumb to their tumour despite intensive re-treatment [9], [10].
Risk stratification is largely based on tumour stage and histology which is key to improving clinical management; 4–10% of WT display anaplasia, which is defined morphologically by the presence of cells with at least threefold nuclear enlargement, hyperchromasia and abnormal mitotic figures [11]. A further distinction is made between focal (FA) and diffuse (DA) anaplasia, based on the topographic distribution of anaplastic elements within the tumour [12]. Presence of DA is the most important adverse prognostic indicator in pre-treated (SIOP) and in chemotherapy-naïve tumours (COG) and patients are assigned to more intensive treatment [13], [14].
TP53 mutations are only found in the anaplastic areas of WT [15], suggesting an association between TP53 and anaplastic cells. Only small numbers of cases of DAWT have been analysed for TP53 mutations, usually as a subgroup of a larger dataset of WT, reflecting the rarity of this histological type. Four studies have clearly analysed 14 DAWT, 12 (86%) displayed mutations in TP53 [15]–[18]. TP53 mutations were also evaluated in anaplastic WT (AWT) without further classification [19]. Although TP53 mutation seems to be predictive of response to treatment and patient survival in several cancers, studies that used immunohistochemistry to investigate p53 prognostic value yielded inconsistent results [20], leading to the conclusion that this technique alone is not suitable for assessing TP53 mutational status. Immunostaining for p53 is positive in ∼76% of anaplastic WT and ∼8% of favourable histology WT - those with ‘nuclear unrest’ which harbour only some of the morphological criteria to be considered anaplastic [21], [22]. In pre-treated patients, p53 positivity was associated to high risk cases that relapsed [23].
Previously, we showed 17p loss in a significant proportion of AWT [24]. Since only small cohorts of DAWT were previously evaluated for TP53 mutation and its clinical implications could therefore not be rigorously assessed, we brought together the largest cohort of anaplastic WT analysed to date to investigate whether the generally accepted frequency of TP53 mutations is correct, and to determine the relationship between TP53 status and patient outcome.
Methods
Patients and samples
Forty patients with unilateral DAWT were included in this analysis. The inclusion criteria were: Wilms tumour with diffuse anaplasia confirmed by central pathology review according to standard international criteria [12] and presence of anaplastic features in the frozen tumour specimen used to extract DNA. Thirty-two cases came from the COG tumour bank and eight from the UK Children's Cancer and Leukaemia tumour bank. All 32 patients from COG were enrolled on the National Wilms Tumor Study-5 protocol (08/1995 to 05/2002) [13] and eight patients were treated in the SIOP WT 2001 trial (02/2002 to 12/2011). All patients were treated according to the high risk arm of their respective trials (Tables S1 and S2).
The frozen tumour samples were taken from a larger cohort of 51 DAWT similarly subject to central pathology review. However 11 tumours were excluded from analyses either because they lacked evidence of anaplasia in the frozen specimen (n = 6) or it was not possible to clearly categorise the haematoxylin-eosin section of the frozen specimen (n = 5).
All samples were obtained from patients whose parent or legal guardian had provided written informed consent for research use (or for older patients, the patient themselves had provided written consent or assent), in accordance with national regulations. Ethical approval for the study was given by East Midlands - Derby Research Ethics Committee (National Research Ethics Service (NRES) in the United Kingdom) with reference approval number MREC/01/4/086 given on 17th January 2002.
Sequencing and analysis of TP53
Genomic DNA was extracted using proteinase K digestion followed by phenol/chloroform extraction as described by manufacture's recommendations. Primer sequences, product lengths and PCR conditions for the 11 exons of TP53 were retrieved from the IARC TP53 website [20]. Sanger sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer's protocol using an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). Sequences were analysed using Mutation Surveyor software v3.20 (SoftGenetics).
Samples negative for TP53 mutations by Sanger sequencing were deep-sequenced to detect mutations present in a minority cell population. Indexed libraries of the PCR products were prepared using the Illumina Nextera XT kit with supplied protocol and sequenced using an Illumina MiSeq instrument, generating 2×150 bp reads with a mean coverage of 162x. Only sequences that passed quality control were analysed further. All sequence alterations were reviewed in the Integrative Genomics Viewer (IGV) version 2.3 [25] and classified as missense, nonsense frameshift, in frame duplication, in frame deletion or splicing mutations by ANNOVAR [26].
Multiplex Ligation-dependent Probe Amplification (MLPA) analysis
Chromosome 17p copy number, where TP53 is located, was assessed by a MLPA assay (P380-X2 custom probemix, MRC-Holland b.v.) according to the manufacturer's protocol (http://www.mrc-holland.com) using 100 ng of DNA (Qubit) with A260/A230 ≥1 (Nanodrop) and containing ≥10 mM Tris-HCl pH 8.2 was used. Data normalisation and analysis was performed with Genemarker v1.85 (Softgenetics).
Analyses of Copy Number and Gene Expression using Microarrays
To infer genomic instability and to confirm 17p loss identified by MLPA, we used genomic copy number BAC arrays from our previously reported data set [24], which has an overlap of 32 patients with the 40 patients from this study. In summary, cases were co-hybridized against unmatched, unrelated pooled normal DNA on the Breakthrough Breast Cancer Research Centre 32k BAC tiling path CGH array. Copy number alterations were calculated based on circular binary segmentation [27], [28] followed by merging of adjacent segments that did not differ significantly in copy number [29], as implemented in the Bioconductor packages DNAcopy and aCGH (http://www.bioconductor.org). Genomic instability was defined in the case of an alternating status in the level of copy number alterations (CNA) [30], [31] using three parameters: 1. More than two contiguous BAC probes with the same copy number, 2. Size range between 1 Kb and 100 Mb to exclude non-focal CNA and 3. Segmentation predicted ≤0.1 for loss and ≥0.1 for gain.
From the same study [24], we retrieved gene expression profiles of 36 cases that also overlap with the 40 patients from this study, and identified differences in gene expression between samples with wild-type and mutated TP53. In summary, tumour and reference cDNA (Universal Human Reference cDNA (Stratagene, La Jolla, CA) were co-hybridized to the Breakthrough Breast Cancer Research Centre 17k 2.1.2 cDNA microarray (ArrayExpress accession number A-MEXP-259). Data are available at GEO (GSE609400).
Statistical analyses
Event-free survival (EFS) was defined to be the time from diagnosis to disease recurrence or other disease related event (including death as a first event from any cause), both clinically identified. Overall survival (OS) analysis included cancer-related deaths, one case where death resulted from infection and one case from toxicity (both had suffered relapse as a first event); the survival interval was calculated relative to the date of diagnosis. Estimates of time-to-event distributions were calculated using the Kaplan-Meier method and differences compared using the log-rank test. Estimates of the relative risk of events between patient subsets were estimated using the Cox regression model. Patients were right censored due to end of study or loss to follow up.
These analyses were performed using IBM SPSS version 21 for Windows and SAS version 9.3. Two-tailed student's t-test assuming unequal variance (given by Levene's test) was used to compare the quantity of copy number alterations between two groups: with wild-type or mutant TP53.
All results were considered significant when p-values were less than 0.05.
Expression analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. The BRB-ArrayTools was used to identify differentially expressed genes between wild-type and mutated TP53, and for gene ontology analyses. After a LOWESS normalization procedure, the class comparison tool uses a Two-sample T-test to find discriminating genes and to confirm their statistical significance p≤0.01. We compared tumours with mutTP53 versus wtTP53. Biological processes were considered over-represented when 1) at least five genes were classified in that process and 2) an observed vs. expected' ratio of >2-fold was observed.
Results
Characterization of TP53 in the tumours
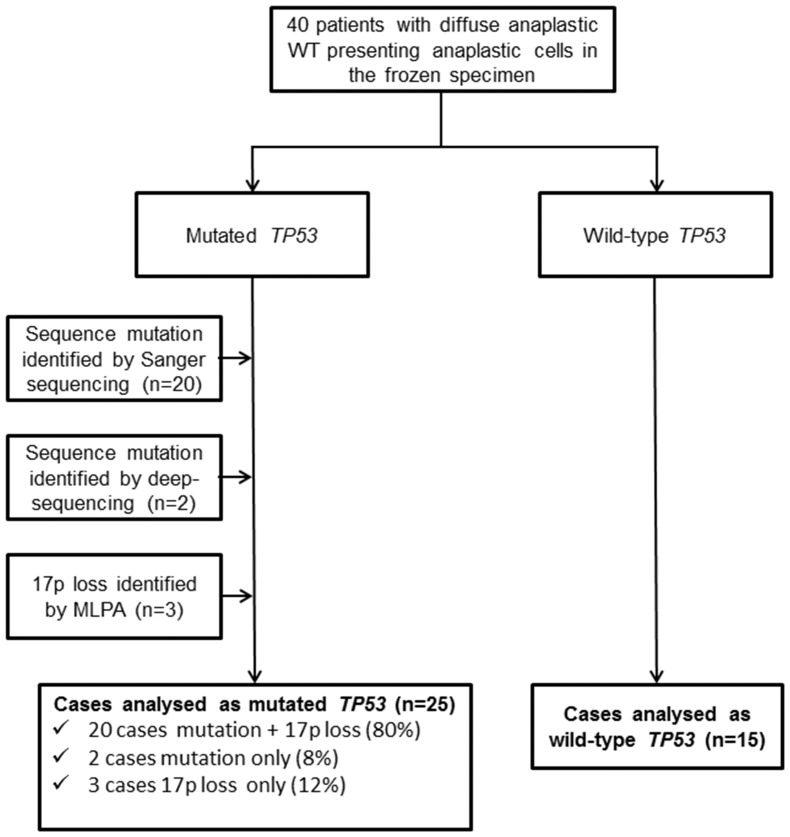
All samples were screened for TP53 mutations in exons 1 to 11, including intron-exon boundaries, with reference to the published wild type sequence (Chr 17 NC_000017.9, Table S3). Distribution of cases with respect to TP53 status is displayed in Figure 1 .
Figure 1. Consort diagram showing the 40 cases of diffuse anaplastic Wilms tumour used for the survival analysis by TP53 mutation and/or 17p loss.
Due to the rarity of DAWT, samples were pooled from two major tumour banks (NWTSG-5 and UK SIOP 2001). There were 51 patients with DA confirmed by central review from which we used 40 for further analyses. For the 40 patients, the median age at diagnosis was 4.6 years (1.42 to 9.17 years, which is older than reported for non-anaplastic WT [32]. Only one patient was aged less than 2 years at diagnosis and did not relapse. The stage distribution was 11 stage I, (27.5%); 8 stage II (20%), 16 stage III (40%); 5 stage IV (12.5%).
Of the 40 cases, 20 (50%) had at least one TP53 mutation detected by Sanger sequencing; one case had two mutations ( Table 1 ). In two further cases, TP53 mutations were only detected by deep sequencing (162-fold). The pathology review noted that DA within the tumour was in the middle of a larger proportion of fat and necrotic cells in one case but the other had a classical presentation of DA. One case (1136) had mutation already described in AWT [16] (Table 1).
Table 1. Cases of diffuse anaplastic WT with TP53 mutations.
| Sample | Trial | Copy number at TP53 locus | Exon | Mutation (annotation in Hg19) |
| Frameshift deletion | ||||
| 1149 | COG | Loss | 10 | g.7573999-7574008het_delTGTTCCGAGA |
| 3036 | COG | Loss | 10 | g.7574000-7574006delTTCCGAG |
| 3048 | COG | Loss | 10 | g.7574013-7574013het_delC |
| Frameshift insertion | ||||
| 3477 | SIOP | Loss | 11 | g.7572933-7572933dupA |
| Nonframeshift deletion | ||||
| 2557 | SIOP | Loss | 5 | g.7578447-7578447dupCATCTACAAGCAGTCACAGCACATGAC |
| 1144 | COG | Loss | 6 | g.7578275-7578277delCTC |
| 1133 | COG | Loss | 10 | g.7574026-7574026dupGGCGTG |
| Nonsynonymous SNV | ||||
| 3044 | COG | Loss | 5 | g.7578406-7578406G>GA¥ |
| 3047 | COG | Loss | 5 | g.7578457-7578457G>GC |
| 3052 | COG | Loss | 5 | g.7578526-7578526G>GT |
| 3056 | COG | Loss | 5 | g.7578413-7578413G>A¥ |
| 3143 | SIOP | Loss | 5 | g.7578394-7578394A>C¥ |
| 1142 | COG | Loss | 8 | g.7577120-7577120G>GA¥ |
| 1706 | SIOP | Loss | 8 | g.7577120-7577120G>GA¥ |
| 2967 | SIOP | Loss | 8 | g.7577129-7577129T>TG |
| 1134† | COG | Loss | 8 | g.7577118-7577118G>GT¥ |
| 1134† | COG | Loss | 8 | g.7577095-7577095C>CA |
| 1145* | COG | Normal | 8 | g.7577124C>T¥ |
| 3041 | COG | Loss | intronic | g.7578298-7578308delCCTCACTGATT |
| 1135 | COG | Normal | intronic | g.7578296-7578305het_delTCACTGATTG |
| 9533 | SIOP | Unknown | 10 | g.7574002C>G¥ |
| Stopgain SNV | ||||
| 4718 | SIOP | Loss | 6 | g.7578212-7578212C>CT¥ |
| 1136 | COG | Loss | 10 | g.7574003-7574003C>T¥ |
| 3145* | SIOP | Loss | 6 | g.7578263-7578263G>A¥ |
SNV: Single Nucleotide Variation.
: Case that had two mutations together with TP53 loss.
*Cases identified by deep-sequencing.
Mutations found in Li Fraumeni patients (IARC TP53 Database, R17).
The other 18 DAWT samples did not have mutations despite clear morphological evidence of anaplasia in the frozen section. All mutations are described in the IARC TP53 Database (http://www-p53.iarc.fr/) [20]. Loss of TP53 was identified by MLPA in an additional 3 cases without TP53 mutation. In total, 20 out of 25 cases (80%) had TP53 mutation and/or 17p loss.
Several additional alterations were previously reported to be normal variations in the population at frequencies of 0.5–60% (source: dbSNP) and assumed to lack functional consequences ( Table 2 ). These variants may have an unknown significance for the patients with AWT, particularly in those three cases with only 17p loss. These were not considered for outcome analysis.
Table 2. Variants with unknown function in cases with 17p loss.
| Cases | SNP location | SNP | dbSNP | MAF |
| Synonymous SNV | ||||
| 3043, 3061, 3864 | Exon 4 | g.7579472-7579472C>G, NM_001126118:c.C98G:p.P33R | rs1042522 | 0.3979 |
| Silent mutations | ||||
| 3043, 3060, 3061 | Exon 11, UTR 3' | g.7572890-7572890A>T | rs1042526 | not available |
| 3043, 3061 | Exon 2, UTR 5' | g.7579801-7579801C>G | rs1642785 | 0.3636 |
| 3043, 3060, 3061 | intronic | g.7579644-7579659del16 | not available | |
MAF: minor allele frequency (source: dbSNP).
Mutations in TP53 effectively risk stratify patients with DAWT
The two cohorts (COG and SIOP) were similar in terms of their clinicopathologic characteristics and overall survival (Tables S1 and S2). Whilst this does not preclude that a larger cohort might display different characteristics, outcomes were similar in both trials. Among 32 patients treated in the COG trial, 13 (41%) relapsed and 11 (34%) died, whereas among 8 patients treated in the SIOP trial, 4 relapsed and died (50%). Typically, patients with DAWT relapse within 2 years and only one patient had a follow-up shorter than 5 years (2.39 years); the median follow-up period of patients still alive (n = 25) was 5.92 years.
For the 17 patients (out of 40) that experienced an event, median time from initial diagnosis to tumour recurrence was 0.75 years (range 0.34–1.61 years) and to death was 1.27 years (range 0.5–2.25 years). We defined mutated cases (mutTP53) as those 25 with damaging aberrations identified by either sequencing methodology, Sanger (20 cases) or deep-sequencing (2 cases), and/or 17p loss identified by MLPA (23 cases) with an overlap of 21 cases between sequence mutations and locus loss. The other 15 cases were considered wild-type TP53 (wtTP53) cases.
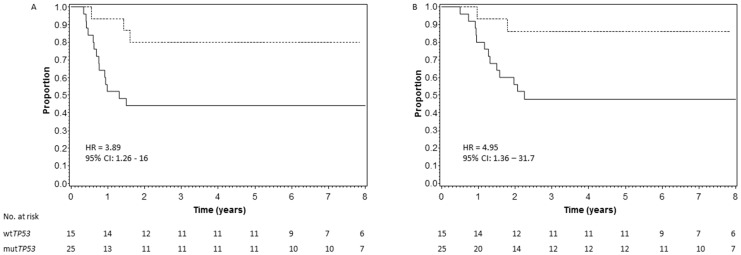
The 15 patients with wtTP53 had statistically significantly better EFS than the 25 patients with mutTP53 (p = 0.02, Figure 2A ). Five-year EFS estimates were 80% for patient with wtTP53 and 44% for patients with mutTP53. The estimated hazard ratio (HR) for failure comparing mutTP53 to wtTP53 was 3.89 (95% profile likelihood confidence interval (CI): 1.26, 16.9).
Figure 2. Patients with diffuse anaplastic WT.
(A) Event-free survival (p = 0.02) and (B) overall survival curves (p = 0.02) for patients with wtTP53 (dashed line) versus mutTP53 (solid line). Numbers at risk are plot every year. HR: hazard ratio, CI: confidence interval.
The results were similar for overall survival. Patients with wtTP53 had statistically significantly better OS than the patients with mutTP53 (p = 0.02, Figure 2B ). Five-year OS estimates were 86% for patients with wtTP53, compared to 48% for patients with mutTP53. The estimated HR for death comparing mutTP53 to wtTP53 was 4.95 (95% profile likelihood CI: 1.36, 31.7). All but one of the 15 deaths was due to tumour progression or relapse. Only a single patient with initial stage I DAWT was known to be alive after relapse (Table S2, case 1136). From the 7 stage I/II patients wild type for TP53 only one relapsed, whilst 6 out of 12 stages I/II patients with mutated TP53 relapsed. Unfortunately, the number of patients used in this study doesn't allow us to draw conclusions regarding the association between stage and relapse or TP53 mutation.
Mutations in TP53 are associated with genomic instability
To determine if TP53 mutation was associated with the genomic instability described previously in tumours with anaplasia, copy number alterations (CNAs) were analysed by aCGH in 32 samples. While cases with wtTP53 (n = 13) had an average of 26.6 (standard deviation (SD) ±18.4) CNAs, cases with mutated TP53 (n = 19) had a significantly higher average of 44.7 (SD ±26.1) CNAs (Table S2). The mean difference was 18.1 (95% CI of difference, 2.0 to 34.2) between groups (p = 0.03, Student's t-test).
Cell cycle pathways are altered in DAWT with TP53 mutations
Gene expression profile was compared between 21 DAWT cases with mutTP53 and 15 DAWT cases with wtTP53. This analysis revealed 93 differentially expressed genes (p≤0.01, t-test, Table S4), with over-representation of genes that participate in cell cycle: AURKA, RNF8, BRCA2, RPS27L, NOL10, RRM2, ORC5, CCNO, HUS1, CDKN1A, FAM83D, PSMC2, NCAPH, PKP4, SUPT5H, USP3, FGFR10P, BUB1, TP53, MLF1, CCNB2, PSMB2, CDC20 and SPC25; and DNA repair processes: RNF8, EGR1, RPS27L, CCNO, HUS1, USP3, TP53, CDKN1A, PSMB2, PSMC2, CCNB2, MORF4L2, TNSF11 and ZMAT3 (Table S5). As expected, TP53 was overexpressed in the wild-type group of samples.
Discussion
This study aimed to identify how frequently TP53 mutation underlies DAWT and whether its presence adds additional prognostic information. In this largest set yet described of DAWT, where molecular analysis was confined to DNA extracted from tissue with confirmed anaplastic changes, we found an overall mutation frequency of only 55% (22/40), lower than previously reported [15]–[18], while 17p loss was observed in 58% (23/40) of the cases, with 20 cases showing both TP53 mutation and 17p loss. Interestingly, from 20 tumours that were TP53 wild-type by Sanger sequencing, mutations were found in further 2 cases by deep sequencing (162-fold). Hence, only 63% (25/40) of definite DAWT had detectable TP53 abnormality even when assessed by the most sensitive technology.
A significant association between poor event free and overall survival and TP53 mutation suggests that it is a potential adverse prognostic factor in addition to anaplastic morphology. The 5-years EFS and OS of patients with DAWT with wtTP53 were 80% and 86%, respectively, not too dissimilar to the reported EFS and OS of patients with favourable histology tumours treated in the same clinical trials [32]–[34]. However these patients receive more intensive treatment, including doxorubicin, with a predictable risk of long term health problems including cardiac failure and second cancer [35], [36]. It remains to be addressed by future studies and clinical trials if patients with DAWT with and without TP53 mutations can be classified into different risk categories and have different treatment strategies.
Ten of the mutations we described have been reported in Li-Fraumeni families ( Table 1 ), an autosomal dominant syndrome associated with germline TP53 mutations, characterised by a high incidence of a range of tumours, with WT not being one of the cardinal tumours included in the diagnostic criteria. Some families harbouring TP53 mutations display a higher WT incidence; five out of six Li-Fraumeni families that developed WT had mutations affecting splicing of TP53 [37], suggesting an association between risk of these patients to develop WT and the type of TP53 mutation. Such mutations account for only 4% of all reported germline mutations [38]. Constitutional DNA was not available to exclude the possibility of these mutations being germline.
In the specific biological context of Li-Fraumeni syndrome and medulloblastoma, TP53 mutations have been associated with chromothripisis, an event characterized by massive genomic rearrangements, causing genomic instability [30], which may be an important evolutionary driver of metastatic ability or chemotherapy treatment resistance [39]. Similar events have been observed in chronic lymphocytic leukaemia [40] and osteosarcomas [41]. We also found that the DAWT with mutated TP53 had more CNAs than the DAWT with wild-type alleles, indicating that the cells are in a state of genomic instability, likely to be a consequence of the mutated TP53. Whether increase in genomic instability is a function of time remains to be addressed.
Hence, unsurprisingly, cell cycle and DNA repair processes were over-represented among the differentially expressed genes. As a cell cycle inhibitory transcription factor, TP53 regulates genes that enforce its growth inhibitory functions in response to oncogenic or damage-induced stress [42]. RPS27L and CDKN1A, known TP53 targets, are down-regulated in tumours without wild-type TP53, a similar finding to a study that evaluated TP53 targets across ten cancer types [43]. Together with TP53, CASP8, CDKN1A, ZMAT3, RRM2 and CCNB2 also belong to the p53 signalling pathway (KEGG), which perturbation is considered a hallmark of cancer [44]. Among these, only CASP8 has been described in WT. CASP8, down-regulated in DAWT with mutated TP53, is a pro-apoptotic factor expressed in favourable histology WT, with no correlation to stage of disease or risk for tumour recurrence [45].
The genes representative of the DNA repair pathway found by this study have not been evaluated in WT. Previous studies reported the lack of defects in the DNA mismatch repair system mainly in favourable histology WT based on the pattern of protein expression of MLH1, PMS2, MSH2 and MSH6 [46], [47] and absence of microsatellite instability [47]. Further work will be necessary to clarify which genes and repair pathways are activated in the mutated TP53 DAWT and at which extension they are important for the other WT subtypes. Analysis of prognostic factors within the histological subgroup of DAWT is challenging, due to its extreme rarity. Less than 10% of Wilms tumour are classified as anaplastic, either focal or diffuse [12]–[14]. Therefore, to obtain a reliable estimate of the frequency of mutated TP53 in DAWT, we made several efforts: 1) Samples were combined through a collaboration between UK and North American clinical trials. 2) In addition to the rigorous central pathology review process of each group, the frozen samples used for DNA extraction were again reviewed to ensure the presence of anaplasia in the specimen, since an association between the visible anaplastic area and TP53 mutation is described [15]. 3) Samples that were negative for mutation by conventional sequencing were re-sequenced using a high-coverage methodology (next-generation sequencing). Altogether, these patients and samples were very carefully reanalysed concerning all aspects of their disease and morphology. Such attention to detail regarding sample selection for mutational analysis will, of course, be required where such an assay may be applied in the real life clinical situation in the future.
TP53 mutational status appears to be an additional indicator of prognosis as it discriminates two classes of anaplastic WT with different outcomes. The value of TP53 mutation analysis in the diagnostic work-up of patients with DAWT should now be tested in an independent cohort in order to answer the important question of whether DAWT without TP53 mutations could be a candidate for reduction in therapy. However, it is possible that the relatively good outcomes were the result of the intensive multi-agent therapy used in modern treatment protocols. As we are approaching the limit of tolerability of classical chemotherapy treatment for these patients, it is clear that novel therapeutic strategies are necessary to cure patients with DAWT and mutTP53.
Supporting Information
Clinical information of the patients.
(PDF)
Detailed clinical and molecular information of the patients with diffuse anaplastic Wilms tumours.
(PDF)
TP53 alterations identified in all diffuse anaplastic Wilms tumours. Alterations were classified as damaging (mutation) using ANNOVAR (http://www.openbioinformatics.org/annovar/).
(PDF)
Differentially expressed genes between AWT with and without TP53 mutation.
(PDF)
Biological Process of the 93 differentially expressed genes. Only classes and parent classes with at least 5 observations in the selected subset and with an 'Observed vs. Expected' ratio of at least 2 genes are shown.
(PDF)
Acknowledgments
The authors thank the CCLG Tissue Bank for access to samples and the investigators of the contributing CCLG centres, including members of the ECMC paediatric network and the COG centres who managed the care of the children entered on clinical and biology studies. We thank UCL Genomics facility for the deep-sequencing experiment.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Cancer Research UK (Grant No. C1188/A11859) and the European Commission Framework Programme 7 P-medicine integrated project (Grant No. FP7-ICT-2009-6) and supported by the National Institute for Health Research Great Ormond Street Hospital UCL Biomedical Research Centre award and Great Ormond Street Hospital for Children's Charitable funds. The Children's Cancer and Leukaemia Group (CCLG) Tissue Bank is funded by Cancer Research UK and CCLG (www.cclg.org.uk). The research was also supported by grants from the National Institutes of Health to the National Wilms Tumor Study Group (CA-42326), the National Wilms Tumor Study Group Late Effects Study (CA-54498), and the Children's Oncology Group (CA-98543 and CA-98413). The funders had no role in study, design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Green DM, Breslow NE, Beckwith JB, Finklestein JZ, Grundy P, et al. (1998) Effect of duration of treatment on treatment outcome and cost of treatment for Wilms' tumor: a report from the National Wilms' Tumor Study Group. J Clin Oncol 16: 3744–3751. [DOI] [PubMed] [Google Scholar]
- 2. Green DM, Breslow NE, Beckwith JB, Ritchey ML, Shamberger RC, et al. (2001) Treatment with nephrectomy only for small, stage I/favorable histology Wilms' tumor: a report from the National Wilms' Tumor Study Group. J Clin Oncol 19: 3719–3724. [DOI] [PubMed] [Google Scholar]
- 3. Breslow NE, Ou SS, Beckwith JB, Haase GM, Kalapurakal JA, et al. (2004) Doxorubicin for favorable histology, Stage II-III Wilms tumor: results from the National Wilms Tumor Studies. Cancer 101: 1072–1080. [DOI] [PubMed] [Google Scholar]
- 4. de Kraker J, Graf N, van Tinteren H, Pein F, Sandstedt B, et al. (2004) Reduction of postoperative chemotherapy in children with stage I intermediate-risk and anaplastic Wilms' tumour (SIOP 93-01 trial): a randomised controlled trial. Lancet 364: 1229–1235. [DOI] [PubMed] [Google Scholar]
- 5. Pritchard-Jones K, Kelsey A, Vujanic G, Imeson J, Hutton C, et al. (2003) Older age is an adverse prognostic factor in stage I, favorable histology Wilms' tumor treated with vincristine monochemotherapy: a study by the United Kingdom Children's Cancer Study Group, Wilm's Tumor Working Group. J Clin Oncol 21: 3269–3275. [DOI] [PubMed] [Google Scholar]
- 6. Green DM (2004) The treatment of stages I-IV favorable histology Wilms' tumor. J Clin Oncol 22: 1366–1372. [DOI] [PubMed] [Google Scholar]
- 7. Pritchard-Jones K, Pieters R, Reaman GH, Hjorth L, Downie P, et al. (2013) Sustaining innovation and improvement in the treatment of childhood cancer: lessons from high-income countries. Lancet Oncol 14: e95–e103. [DOI] [PubMed] [Google Scholar]
- 8. Mitchell C, Pritchard-Jones K, Shannon R, Hutton C, Stevens S, et al. (2006) Immediate nephrectomy versus preoperative chemotherapy in the management of non-metastatic Wilms' tumour: results of a randomised trial (UKW3) by the UK Children's Cancer Study Group. Eur J Cancer 42: 2554–2562. [DOI] [PubMed] [Google Scholar]
- 9. Dome JS, Liu T, Krasin M, Lott L, Shearer P, et al. (2002) Improved survival for patients with recurrent Wilms tumor: the experience at St. Jude Children's Research Hospital. J Pediatr Hematol Oncol 24: 192–198. [DOI] [PubMed] [Google Scholar]
- 10. Malogolowkin M, Cotton CA, Green DM, Breslow NE, Perlman E, et al. (2008) Treatment of Wilms tumor relapsing after initial treatment with vincristine, actinomycin D, and doxorubicin. A report from the National Wilms Tumor Study Group. Pediatr Blood Cancer 50: 236–241. [DOI] [PubMed] [Google Scholar]
- 11. Beckwith JB, Palmer NF (1978) Histopathology and prognosis of Wilms tumors: results from the First National Wilms' Tumor Study. Cancer 41: 1937–1948. [DOI] [PubMed] [Google Scholar]
- 12. Faria P, Beckwith JB, Mishra K, Zuppan C, Weeks DA, et al. (1996) Focal versus diffuse anaplasia in Wilms tumor–new definitions with prognostic significance: a report from the National Wilms Tumor Study Group. Am J Surg Pathol 20: 909–920. [DOI] [PubMed] [Google Scholar]
- 13. Dome JS, Cotton CA, Perlman EJ, Breslow NE, Kalapurakal JA, et al. (2006) Treatment of anaplastic histology Wilms' tumor: results from the fifth National Wilms' Tumor Study. J Clin Oncol 24: 2352–2358. [DOI] [PubMed] [Google Scholar]
- 14. Reinhard H, Schmidt A, Furtwangler R, Leuschner I, Rube C, et al. (2008) Outcome of relapses of nephroblastoma in patients registered in the SIOP/GPOH trials and studies. Oncol Rep 20: 463–467. [PubMed] [Google Scholar]
- 15. Bardeesy N, Beckwith JB, Pelletier J (1995) Clonal expansion and attenuated apoptosis in Wilms' tumors are associated with p53 gene mutations. Cancer Res 55: 215–219. [PubMed] [Google Scholar]
- 16. Bardeesy N, Falkoff D, Petruzzi MJ, Nowak N, Zabel B, et al. (1994) Anaplastic Wilms' tumour, a subtype displaying poor prognosis, harbours p53 gene mutations. Nat Genet 7: 91–97. [DOI] [PubMed] [Google Scholar]
- 17. el Bahtimi R, Hazen-Martin DJ, Re GG, Willingham MC, Garvin AJ (1996) Immunophenotype, mRNA expression, and gene structure of p53 in Wilms' tumors. Mod Pathol 9: 238–244. [PubMed] [Google Scholar]
- 18. Malkin D, Sexsmith E, Yeger H, Williams BR, Coppes MJ (1994) Mutations of the p53 tumor suppressor gene occur infrequently in Wilms' tumor. Cancer Res 54: 2077–2079. [PubMed] [Google Scholar]
- 19. Takeuchi S, Bartram CR, Ludwig R, Royer-Pokora B, Schneider S, et al. (1995) Mutations of p53 in Wilms' tumors. Mod Pathol 8: 483–487. [PubMed] [Google Scholar]
- 20. Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, et al. (2007) Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629. [DOI] [PubMed] [Google Scholar]
- 21. Hill DA, Shear TD, Liu T, Billups CA, Singh PK, et al. (2003) Clinical and biologic significance of nuclear unrest in Wilms tumor. Cancer 97: 2318–2326. [DOI] [PubMed] [Google Scholar]
- 22. Jadali F, Sayadpour D, Rakhshan M, Karimi A, Rouzrokh M, et al. (2011) Immunohistochemical detection of p53 protein expression as a prognostic factor in Wilms tumor. Iran J Kidney Dis 5: 149–153. [PubMed] [Google Scholar]
- 23. Franken J, Lerut E, Van Poppel H, Bogaert G (2013) p53 Immunohistochemistry Expression in Wilms Tumor: A Prognostic Tool in the Detection of Tumor Aggressiveness. J Urol 189: 664–670. [DOI] [PubMed] [Google Scholar]
- 24. Williams RD, Al-Saadi R, Natrajan R, Mackay A, Chagtai T, et al. (2011) Molecular profiling reveals frequent gain of MYCN and anaplasia-specific loss of 4q and 14q in Wilms tumor. Genes Chromosomes Cancer 50: 982–995. [DOI] [PubMed] [Google Scholar]
- 25. Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olshen AB, Venkatraman ES, Lucito R, Wigler M (2004) Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572. [DOI] [PubMed] [Google Scholar]
- 28. Venkatraman ES, Olshen AB (2007) A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23: 657–663. [DOI] [PubMed] [Google Scholar]
- 29. Willenbrock H, Fridlyand J (2005) A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics 21: 4084–4091. [DOI] [PubMed] [Google Scholar]
- 30. Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, et al. (2012) Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, et al. (2009) Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet 41: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pritchard-Jones K, Moroz V, Vujanic G, Powis M, Walker J, et al. (2012) Treatment and outcome of Wilms' tumour patients: an analysis of all cases registered in the UKW3 trial. Ann Oncol 23: 2457–2463. [DOI] [PubMed] [Google Scholar]
- 33. Dome JS, Fernandez CV, Mullen EA, Kalapurakal JA, Geller JI, et al. (2013) Children's Oncology Group's 2013 blueprint for research: renal tumors. Pediatr Blood Cancer 60: 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smets AM, van Tinteren H, Bergeron C, De Camargo B, Graf N, et al. (2012) The contribution of chest CT-scan at diagnosis in children with unilateral Wilms' tumour. Results of the SIOP 2001 study. Eur J Cancer 48: 1060–1065. [DOI] [PubMed] [Google Scholar]
- 35. Breslow NE, Lange JM, Friedman DL, Green DM, Hawkins MM, et al. (2010) Secondary malignant neoplasms after Wilms tumor: an international collaborative study. Int J Cancer 127: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Termuhlen AM, Tersak JM, Liu Q, Yasui Y, Stovall M, et al. (2011) Twenty-five year follow-up of childhood Wilms tumor: a report from the Childhood Cancer Survivor Study. Pediatr Blood Cancer 57: 1210–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scott RH, Stiller CA, Walker L, Rahman N (2006) Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet 43: 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, et al. (2002) The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat 19: 607–614. [DOI] [PubMed] [Google Scholar]
- 39. Hanel W, Moll UM (2012) Links between mutant p53 and genomic instability. J Cell Biochem 113: 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zenz T, Krober A, Scherer K, Habe S, Buhler A, et al. (2008) Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 112: 3322–3329. [DOI] [PubMed] [Google Scholar]
- 41. Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, et al. (2003) The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci U S A 100: 11547–11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412. [DOI] [PubMed] [Google Scholar]
- 43. Parikh N, Hilsenbeck S, Creighton CJ, Dayaram T, Shuck R, et al. (2013) Effects of TP53 Mutational Status on Gene Expression Patterns Across Ten Human Cancer Types. J Pathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfeifer GP, Hainaut P (2011) Next-generation sequencing: emerging lessons on the origins of human cancer. Curr Opin Oncol 23: 62–68. [DOI] [PubMed] [Google Scholar]
- 45. Miller MA, Karacay B, Breslow NE, Li S, O'Dorisio MS, et al. (2005) Prognostic value of quantifying apoptosis factor expression in favorable histology wilms tumors. J Pediatr Hematol Oncol 27: 11–14. [DOI] [PubMed] [Google Scholar]
- 46. Segers H, van den Heuvel-Eibrink MM, de Krijger RR, Pieters R, Wagner A, et al. (2012) Defects in the DNA mismatch repair system do not contribute to the development of childhood Wilms tumors. Pediatr Dev Pathol. [DOI] [PubMed] [Google Scholar]
- 47. Diniz G, Aktas S, Cubuk C, Ortac R, Vergin C, et al. (2013) Tissue expression of MLH1, PMS2, MSH2, and MSH6 proteins and prognostic value of microsatellite instability in Wilms tumor: experience of 45 cases. Pediatr Hematol Oncol 30: 273–284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical information of the patients.
(PDF)
Detailed clinical and molecular information of the patients with diffuse anaplastic Wilms tumours.
(PDF)
TP53 alterations identified in all diffuse anaplastic Wilms tumours. Alterations were classified as damaging (mutation) using ANNOVAR (http://www.openbioinformatics.org/annovar/).
(PDF)
Differentially expressed genes between AWT with and without TP53 mutation.
(PDF)
Biological Process of the 93 differentially expressed genes. Only classes and parent classes with at least 5 observations in the selected subset and with an 'Observed vs. Expected' ratio of at least 2 genes are shown.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.