Figure 5.
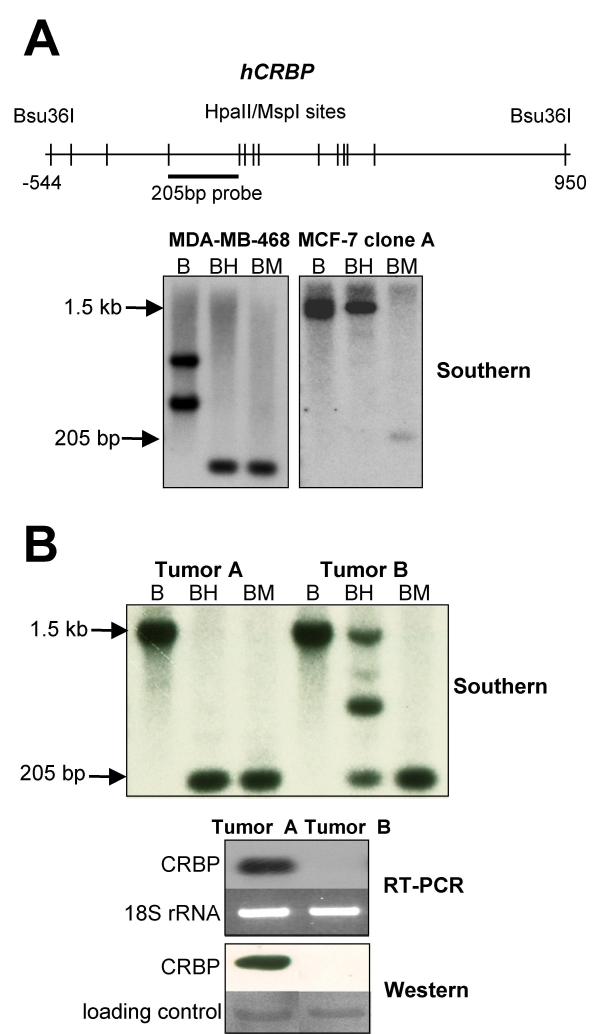
In some human breast cancers, hCRBP hypermethylation is widespread. A. Sketch outlining the Southern blot strategy used to evaluate hCRBP methylation in human breast cancer specimens. The strategy was tested in MDA-MB-468 and ZR75-1 cells whose hCRBP methylation status was known beforehand. B, BH and BM stand for, respectively, Bsu36I digestion alone, Bsu36I digestion followed by Hpall digestion, and Bsu36I digestion followed by MspI digestion (MspI is the methylation-insensitive isoschizomer of Hpall). In agreement with the bisulfite sequencing data, Southern blotting revealed hCRBP undermethylation in MDA-MB-468 and hypermethylation in MCF-7 clone A cells (note loss of the large Bsu36I fragment after BH digestion of MDA-MB-468 genomic DNA and retention of this fragment in the case of MCF-7 clone A). A hCRBP restriction fragment length polymorphism (an extra Bsu36I site within the 205 bp fragment) accounts for the displaced band migration pattern in MDA-MB-468 cells. B. Southern, RT-PCR and Western blotting results for 2 human breast cancer specimens. The yield of the 205 bp product was decreased by >50% after BH digestion relative to BM digestion. We infer from this that >50% of the cells in the tumor displayed hCRBP hypermethylation. The protein blot was post-stained with Ponceau S and a major band used as loading control.