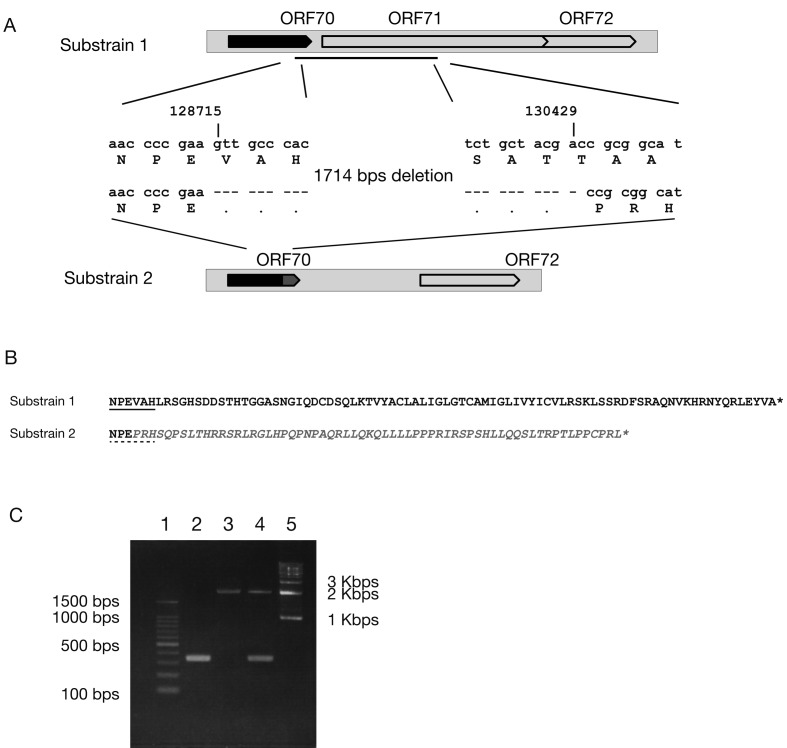
Fig. 2.
(A) Nucleotide deletion from the tail part of ORF70 to the head part of ORF71. The arrow in black indicates the original ORF70. The other arrow in dark grey indicates the truncated tail part of ORF70 caused by deletion. (B) Amino acid sequence alignments of the tail part of ORF70 in Substrains 1 and 2. The amino acid sequences with underline indicate the corresponding sequences shown in the panel A. The amino acid sequence in italic grey indicates the truncated tail of ORF70 caused by the deletion. The asterisks (*) indicate the placement of stop codon in the original nucleotide sequence. (C) PCR results to confirm the presence of 2 viruses in T-616. PCR primers used were as follows: Zebra_71-F 5′-ccaacgtaccatcaagtgcggta-3′ and Zebra_71-R 5′-cgctggtactctcgtaggttgac-3′. PCR was examined by using PrimeSTAR Max Premix (TaKaRa Bio, Otsu, Japan) with amplification program as follows: the primary hold at 95°C for 4 min, 30 cycles of 98°C 10 sec, 55°C 15 sec and 72°C 45 sec. Lanes were 100 bp ladder marker (1), T-616 substrain 2 (2), T-616 substrain 1 (3), the original seed stock of T-616 (4) and 1 kbp ladder marker (5). Expected sizes are 344 bps for T-616 substrain 2 and 2,058 bps for T-616 substrain 1.