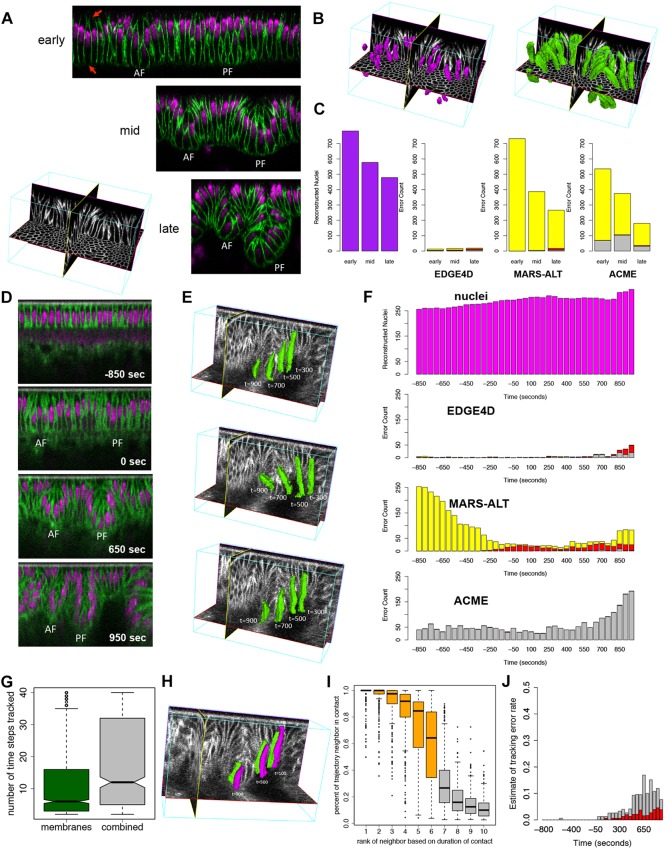
Fig. 1.
Assessment of cell shape reconstruction and tracking. (A) Mid-sagittal slices of the dorsal folds from fixed Drosophila embryos staged at early, mid and late stages. AF, anterior fold; PF, posterior fold. Membranes were labeled by antibody to Neurotactin and nuclei by OliGreen DNA dye. The red arrows highlight missing membrane signal along the basal and apical ends of the cells. (B) Examples of a subset of nuclei (purple) and cell shapes (green) reconstructed from the mid-stage fixed data set. Based on their nuclei, virtually all cells could be correctly identified, providing a ground truth for testing their identification based on membrane labeling alone. (C) Quantification of the number of nuclei (purple) at each stage. The greater number of nuclei at the early time point is due to the larger field of view. Error counts are shown for EDGE4D, MARS-ALT and ACME applied to only the membrane channel in the data sets. Merged cells (red), broken cells (gray) and missed cells (yellow) were detected using the nuclei. (D) Mid-sagittal slices of the dorsal folds from a live data set collected using two-photon imaging. Time 0 s designates the completion of cellularization. Membranes were labeled with Resille-GFP and nuclei were labeled with H2Av-mRFP. (E) Examples of three different cells tracked into the posterior fold. Cell shapes are shown for time points 300 s, 500 s, 700 s and 900 s past completion of cellularization in the live two-photon data set. (F) Segmentation error counts across the time points collected. Merged cells (red), broken cells (gray) and missed cells (yellow) were detected using the nuclei reconstructions as ground truth. (G) The number of time points that a cell was tracked using the membrane channel alone (green) or the combined membrane and nuclear signal (gray). (H) Example of two cells that maintain stable contact as they descend into the posterior fold at 100 s, 500 s and 900 s past the completion of cellularization. (I) Analysis of neighbor contact stability for 211 cells tracked for more than ten time points. (G,I) The ends of the boxes are the quartiles; the centerline is the median; notches, where present, provide the 95% confidence interval around the median; and whiskers extend 1.5 times the interquartile range. (J) Assessment of the tracking error rate based on the number of events in which more than half the neighbors of a cell changed for cells tracked ten time points or more using membrane signal only (gray) or membrane and nuclei (red).