Figure 1.
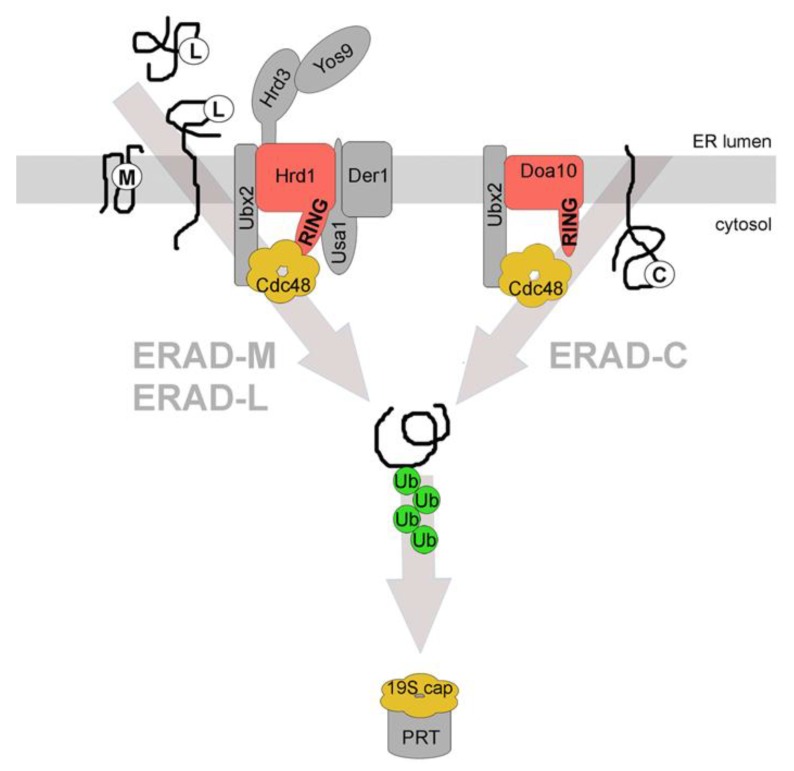
Distinct endoplasmic reticulum-associated degradation (ERAD) pathways. Three main ERAD pathways in yeast are classified based on substrates and the components that are involved in their degradation. ERAD-L degrades membrane integrated or soluble proteins with misfolded domains in the ER lumen, marked with (L). All depicted constituents of the Hrd1-complex are required for the efficient degradation of these substrates. ERAD-M degrades membrane integrated proteins with misfolded regions in their transmembrane domain(s), marked with (M). Proteins of this class are degraded via the Hrd1-complex but do not require Usa1p and Der1p for efficient degradation. ERAD-C degrades membrane integrated proteins with misfolded domains in the cytoplasm, marked with (C). These proteins are degraded via the Doa10-complex. All ERAD pathways require cytosolic Cdc48p for substrate retrotranslocation and extraction from the ER membrane. The substrate is ubiquitinated by the E3 ligases during or after retrotranslocation and is targeted to the proteasome (PRT) for degradation. Cdc48p and the 19S cap of the proteasome have structural and functional similarities. The classification for the different ERAD pathways also exists in mammalian cells albeit it is less stringent. RING = RING domain of the E3 ligases that are shown in red. The associated constituents of the individual complexes are shown in grey. Ub = ubiquitin. See text for details.