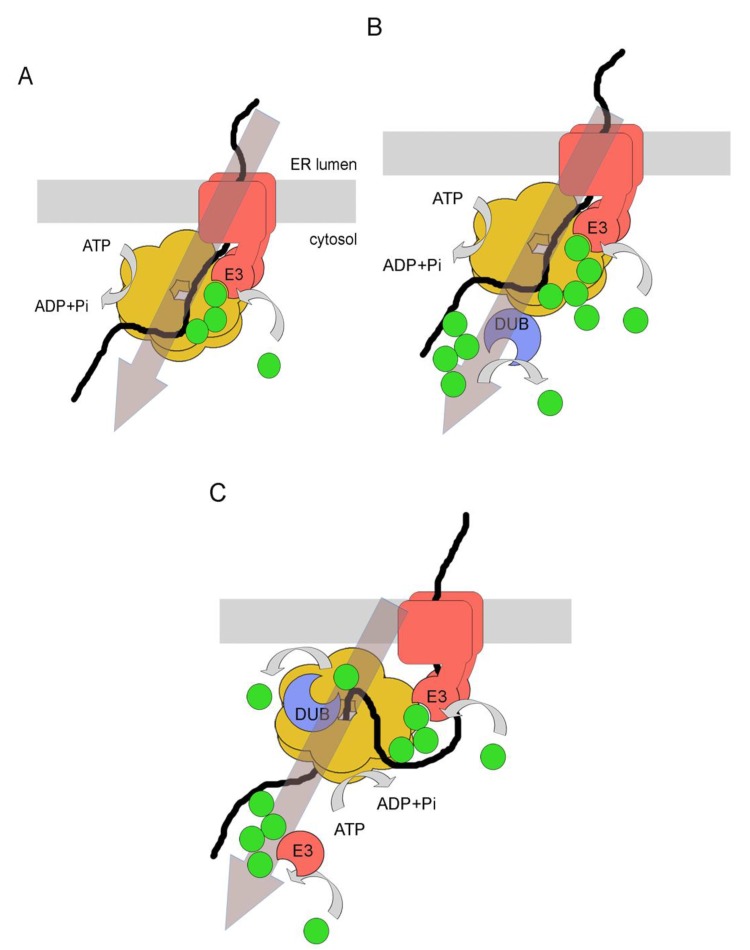
Figure 3.
Models for PUC processing as a mechanism for protein retrotranslocation. (A) Ubiquitination of the substrate on the cytosolic side of the ER membrane by an E3 ligase promotes binding to the hexameric Cdc48p, via its ubiquitin-binding cofactors Npl4p and Ufd1p. ATP hydrolysis by Cdc48p is thought to promote the complete retrotranslocation and substrate extraction from the ER membrane. See text for details. (B) In the mammalian system, the p97-associated DUB Ataxin-3 acts downstream of protein retrotranslocation and might promote the removal of ubiquitins from longer PUCs. This is thought to be required for the generation of PUCs with optimal binding affinities to proteasomal shuttle factors or to the proteasome itself. See text for details. (C) The p97-associated DUB YOD1 is thought to act upstream of protein retrotranslocation. Its activity was suggested to be needed for the complete removal of PUCs to allow passage of the substrate through the pore of p97. ATP hydrolysis would then couple protein retrotranslocation with protein unfolding. Subsequently, reubiquitination by an E3 ligase would allow substrate targeting to the proteasome. See text for details.