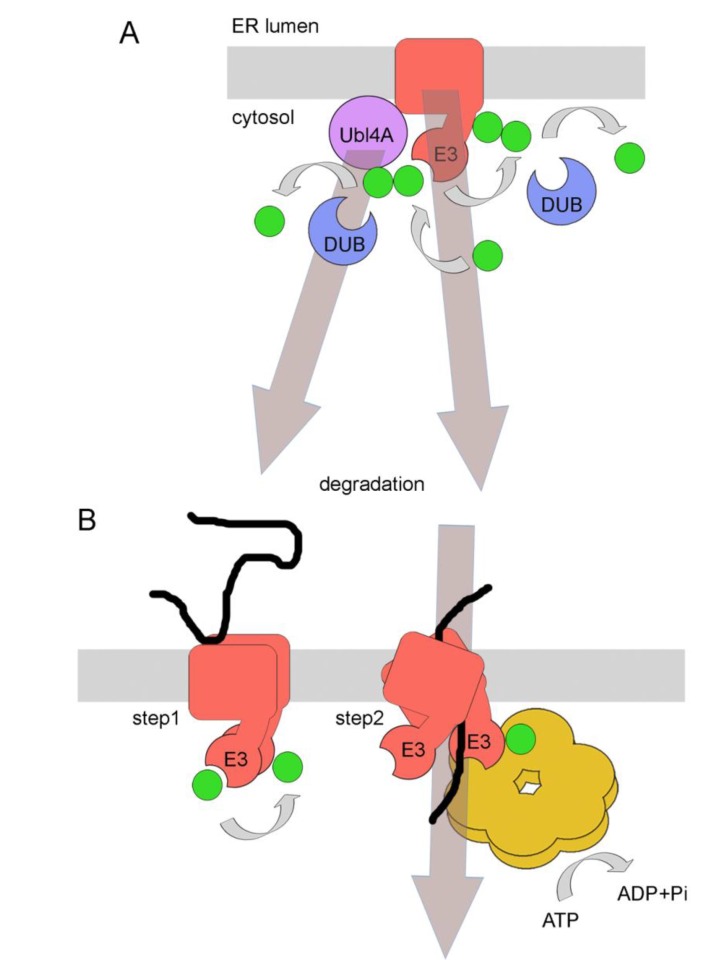
Figure 5.
Models for ubiquitination and deubiquitination of ERAD machinery components as a mechanism to regulate ERAD. (A) Regulation through degradation. In the mammalian system, ubiquitination of the ERAD component Ubl4A by the E3 ligase gp78 leads to the degradation of the associated shuttling chaperone Bag6, thereby reducing efficient substrate targeting to the proteasome. Ubl4A ubiquitination is reversed by the DUB Usp13. A similar mechanism involves the self-ubiquitination of Hrd1p in yeast in absence of Hrd3p and Usa1p, leading to effective degradation of the E3 ligase. No DUB is currently known in this system. See text for details. (B) Regulation through conformational changes. Binding of a substrate to Hrd1p in the ER lumen induces oligomerization and self-ubiquitination of the E3 ligase (step 1). Subsequently, ATP hydrolysis by bound Cdc48p would result in conformational changes in the ATPase, which will consequently result in conformational changes in Hrd1p oligomers. This, in turn, would push the substrate through a postulated channel formed by Hrd1p oligomers (together with associated components like Der1p) until it is partially exposed to the cytosol (step 2). See text for details.