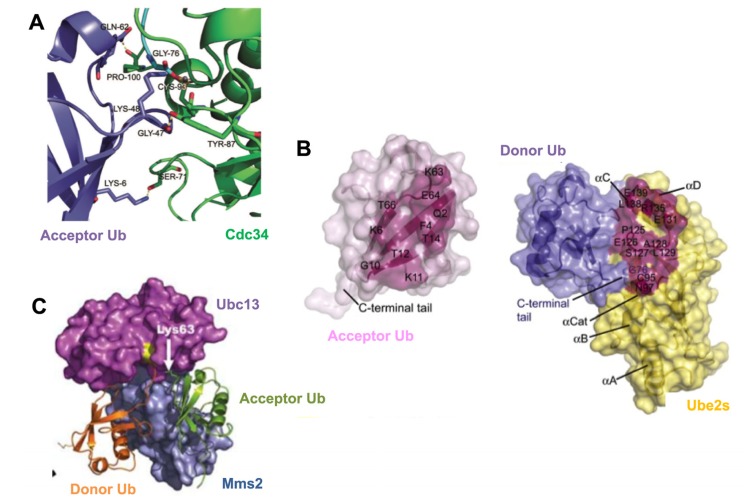
Figure 2.
Mechanisms for generating lysine-specific poly-Ub chains.
(A) Computational docking of the human E2, Cdc34 (green), in complex with an acceptor Ub (purple). Serine-71, tyrosine-87 and proline-100 on human Cdc34 interact with lysine-6, glycine-47 and glutamine-62 on acceptor Ub, respectively, to align K48 of acceptor Ub in the Cdc34 catalytic site and promote K48-linked poly-Ub chain formation [33]. (B) Surface representation of an acceptor Ub (pink, left) and the donor Ub (blue) in complex with Ube2s (yellow, right), based on computational docking and mutational analysis. The residues that make intermolecular contacts are highlighted, which allow optimal positioning of acceptor Ub K11 toward the Ube2S~Ubdonor thioester bond to catalyze K11-specific poly-Ub chains [36]. Copyright (2011), with permission from Elsevier (C) Crystal structure of Mms2 (blue) and Ubc13 (magenta) in association with acceptor Ub (green) and a donor Ub (orange). Acceptor Ub is bound by Mms2 and Ubc13 to position K63 toward the Ubc13~Ubdonor thioester bond [38]. Copyright (2006), with permission from Nature Publishing Group.