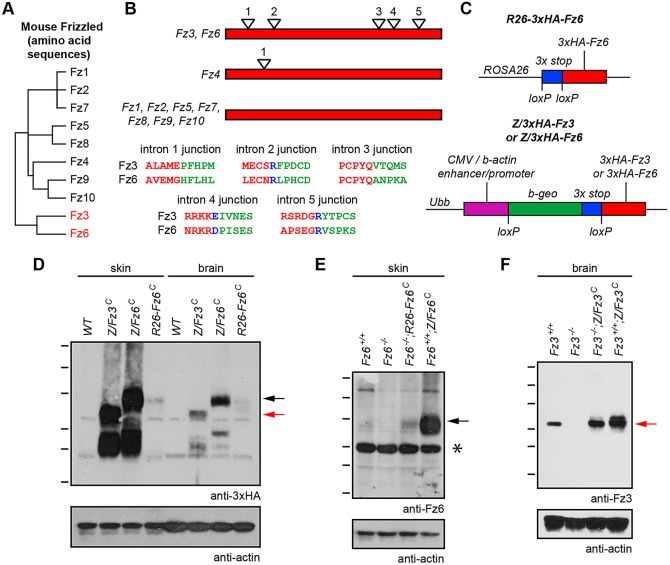
Fig. 1.
Knock-in alleles for constitutive production of Fz3 and Fz6. (A) Dendrogram showing amino acid sequence identities among the 10 mouse Fz proteins. Fz3 and Fz6 show 48% amino acid identity. (B) Schematic of coding region intron-exon structures of mouse Fz family members. Fz3 and Fz6 each have five introns, and the intron positions are precisely conserved, as seen by the alignment of encoded amino acids near each exon-exon junction. Red lettering: amino acids encoded by the 5′ exon. Green lettering: amino acids encoded by the 3′ exon. Blue lettering: the intron is located within that codon. Fz4 has one coding region intron; all other genes in the Fz family lack coding region introns. (C) Schematic of ROSA26-3xHA-Fz6 (top), and Z/3xHA-Fz3 and Z/3xHA-Fz6 (bottom). At the Z locus, Cre-mediated deletion of the loxP-beta-geo-stop-loxP cassette leads to constitutive expression of 3xHA-Fz3 or 3xHA-Fz6 driven by the CAG promoter. At the ROSA26 (R26) locus, Cre-mediated deletion of the loxP-stop-loxP cassette leads to constitutive expression of 3xHA-Fz6 driven by the relatively weak ROSA26 promoter. The constitutively active derivatives of these alleles are referred to as Z/Fz3C, Z/Fz6C and R26-Fz6C, respectively. (D-F) Anti-3xHA, anti-Fz6 and anti-Fz3 immunoblots of P1 brain and skin extracts from wild-type, Z/Fz3C, Z/Fz6C and R26-Fz6C mice in the presence or absence of endogenous Fz3 or Fz6 alleles, as indicated. The Fz3−/− brain was harvested at E18.5. The ubiquitously expressed (D) and endogenous (E) Fz6 proteins migrate at higher apparent molecular weights than the ubiquitously expressed (D) and endogenous (F) Fz3 proteins. Molecular weight (MW) heterogeneity may reflect heterogenous glycosylation. Black arrows, Fz6 protein; red arrows, Fz3 protein. Asterisk indicates an irrelevant cross-reacting band. MW standards are 180, 115, 82, 64, 49 and 37 kDa.