Figure 1.
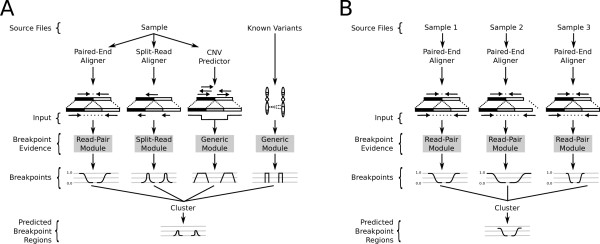
The LUMPY framework for integrating multiple structural variation signals. (A) A scenario in which LUMPY integrates three different sequence alignment signals (read-pair, split-read and read-depth) from a genome single sample. Additionally, sites of known variants are provided to LUMPY as prior knowledge in order to improve sensitivity. (B) A single signal type (in this case, read-pair) that is integrated from three different genome samples. We present these as example scenarios and emphasize that multi-signal and multi-sample workflows are not mutually exclusive. CNV, copy number variation.
