Figure 1.
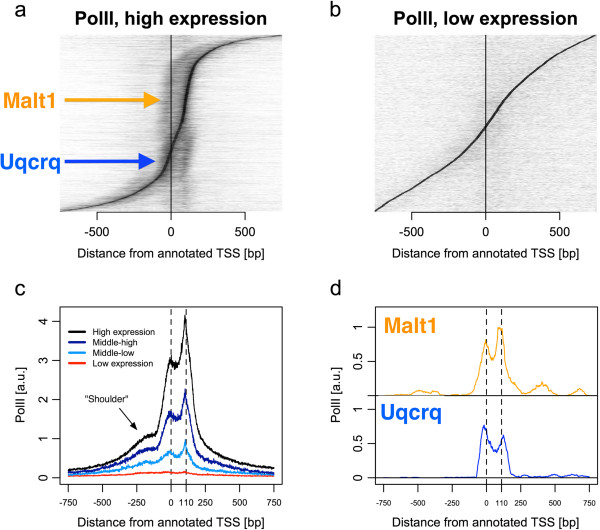
High-resolution PolII profiles show distinct accumulation peaks at the TSS and + 110 ± 20 bp downstream. Sorted ChIP-seq profiles and genome-averaged profiles of normalized PolII signal on promoters are displayed from 5′ to 3′ centered at each TSS. Transcripts are separated by microarray expression into lowly expressed (<6.0 microarray units), moderately expressed (between 6.0 and 8.0 units) and highly expressed (>8.0 units) (see also Figure 2b). (a, b) Stacked profiles, sorted vertically by position of maximum signal, with signal strength normalized to give the same shade of grey at the maximum for each profile. (c) Genome-averaged profile of PolII occupancy for highly expressed (black), moderately expressed divided into two quantiles (two shades of blue) and lowly expressed (red) transcripts. The two dashed lines indicate the two main peaks at TSS and TSS + 110. (d) Examples of genes with different peak-height ratios, not normalized to their maxima: Malt1 (orange, peak-height ratio 1.28) and Uqcrq (blue, peak-height ratio 0.93) as indicated in (a). a.u., arbitrary units.
