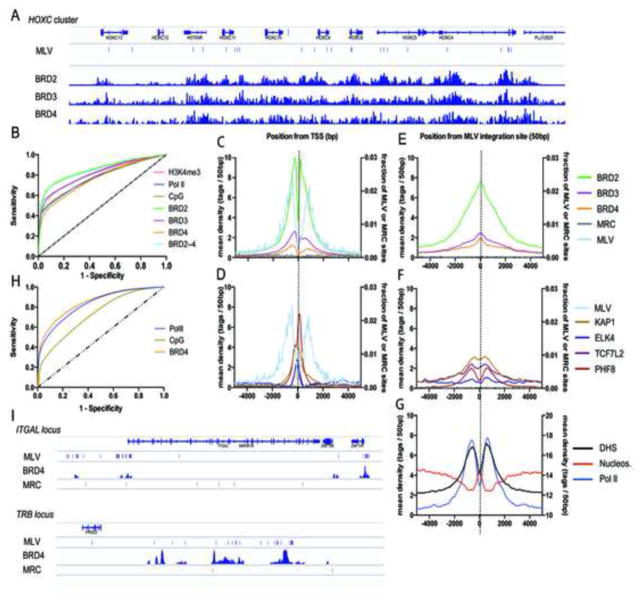
Figure 3. MLV vectors integrate in BET protein hot spots.
A) Schematic representation of the BET chromatin binding profile (Leroy et al., 2012) and MLV integration sites (MLV) in the HOXC cluster in 293T cells. Matched random control sites (MRC) were absent in this region. B) Receiver-operator characteristic (ROC) analysis of BRD2–4, Pol II, H3K4me3 and CpG islands (Berry et al., 2006). The area under the curve (AUC) is calculated for the different markers and shown in Supplementary Table 3. (C–G) Mean background-subtracted sequencing read density in 50 bp bins in a 10 kb window around (C, D) transcription start sites (TSSs) or (E–G) MLV integration sites for (C, E) BRD2–4 (ChIP-Seq), (D, F) the unrelated transcription factors PHF8, ELK4, KAP1 and TCF7L2 (ChIP-Seq) or (G) open chromatin (DNase I Hypersensitivity, DHS), Pol II (ChIP-Seq) and nucleosome positions (Micrococcal Nuclease-Seq, MN-Seq) plotted on the right y-axis. In (C–F) the number of MLV and MRC sites is plotted on the right y-axis as a fraction of the total number of respective sites. (H) ROC analysis of BRD4, Pol II and CpG islands in human primary CD4+ T-cells. Corresponding AUC values are given in Supplementary Table 3. (I) Schematic representation of the ITGAL (Integrin alpha-L) locus and the 3′ end of the TRB (T-cell receptor beta) locus highlighting MLV integration sites, BRD4 peaks and MRC sites.