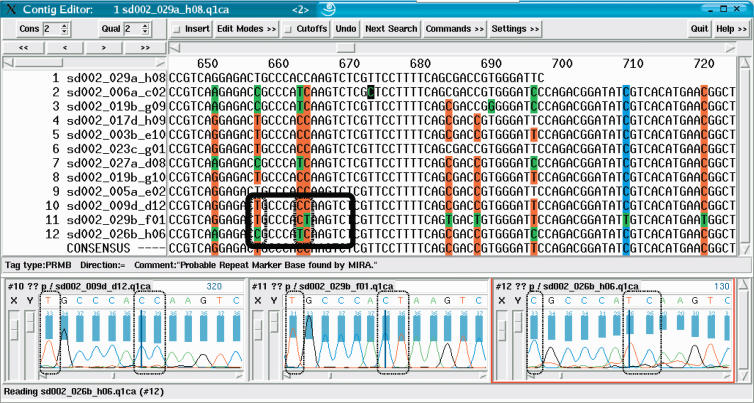
Figure 4.
Snapshot of a contig in the sequence assembly after the first iteration (visual representation by means of the gap4 program). All sequences were assembled together. After the assembly, miraEST searched for unresolved mismatches with good signal qualities, tagging entire columns as dangerous potential SNP sites for the next iteration. miraEST tagged strong SNP sites bright red, weak sites in blue; bases differing from the consensus are shown in green by the gap4 program. Some bases were not tagged, although they cover a possible SNP site; these bases generally have trace signals of bad quality that the assembler deemed to be too dangerous to be taken as differentiation criterion. miraEST will dismantle that contig and reassemble the sequences immediately, this time using the information gained about the potential SNP sites in the previous assembly to correctly discern between different mRNA transcripts having different SNP variants. The black rectangle amidst the sequences depicts the three trace signal extracts that have been exemplarily shown below; the smaller black boxes within the rectangle depict the discrepancy bases that have also been surrounded by black boxes in the traces. All sequences have indisputable trace curves and quality values (shown as a blue bar above the traces). One can clearly see that there will be at least three different mRNA transcripts to be built, on the basis of the double-base mutation in the middle of the box, one reading CC, the next CT, and the last TC.