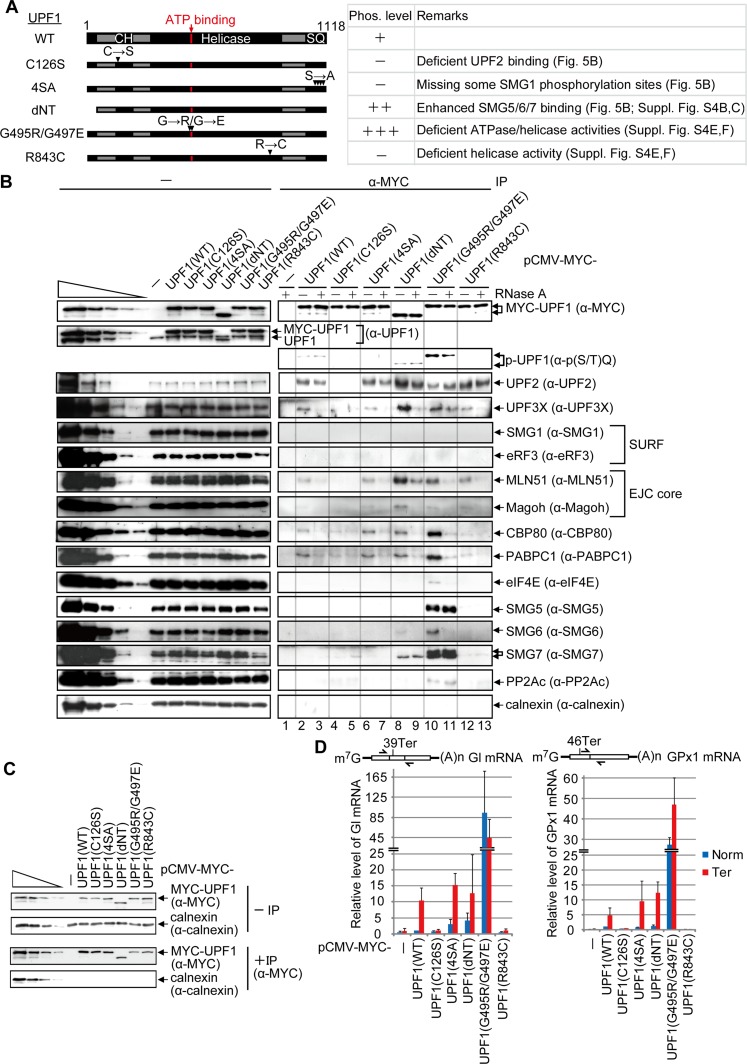
Figure 5.
UPF1 recognition of NMD targets requires ATPase and helicase activities. (A) Diagrams of UPF1 variants denoting the CH region and the region containing the four serine–glutamine (SQ) SMG1 phosphorylation sites. Horizontal arrows denote amino acid changes at positions specified by downward-facing arrowheads. (B) HEK293T cells (8 × 107 per 150-mm dish) were transiently transfected with 0.6 μg of pmCMV-MYC-UPF1(WT), 2 μg of MYC-UPF1(C126S), 1 μg of MYC-UPF1(4SA), 0.5 μg of MYC-UPF1(dNT), 4 μg of MYC-UPF1(G495R/G497E), 0.8 μg of MYC-UPF1(R843C), or, as a negative control, 0.5 μg of pmCMV-MYC. Cell lysates were treated with (+) or without (−) RNase A and subsequently immunoprecipitated using anti-MYC (α-MYC). Western blotting before (−) or after (α-MYC) immunoprecipitation. For samples after immunoprecipitation, fivefold less immunoprecipitate was loaded in the analysis of MYC-UPF1 compared with the analysis of other proteins. (C) HEK293T cells were transiently transfected as described in B except that transfections included 0.5 μg of phCMV-MUP and 1 μg each of pmCMV-Gl and pmCMV-GPx1, either Norm or Ter. Cell lysates were immunoprecipitated using anti-MYC and analyzed by Western blotting. (D) RT-qPCR of full-length Gl or GPx1 mRNA from samples in C. The level of Gl or GPx1 mRNA before immunoprecipitation was normalized to the level of MUP mRNA, and, given the comparable immunoprecipitation efficiencies, the normalized level of Gl Norm or GPx1 Norm mRNA after immunoprecipitation that coimmunoprecipitated with MYC-UPF1(WT) is defined as 1. Diagrams are of Gl mRNA or GPx1 mRNA as in Figure 2A, and the arrows specify PCR primer positions. Quantitations derive from two to three independently performed experiments and represent the mean plus standard deviations.