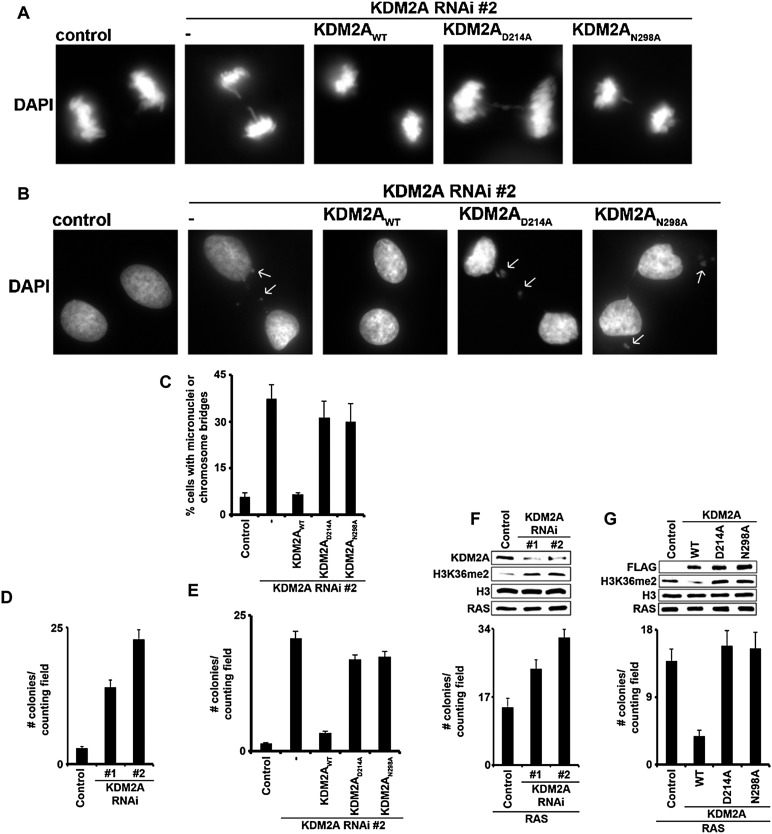
Figure 5.
Lysine demethylase activity of KDM2A is essential for the maintenance of genomic stability and the suppression of cellular transformation. (A) Loss of KDM2A catalytic activity leads to chromosome missegregation during mitosis. Representative images of 4′,6-diamidino-2-phenylindole (DAPI) staining of mitotic HT1080 cells carrying control vector or stably expressing KDM2AWT, KDM2AD214A, or KDM2AN298A. (B,C) KDM2A catalytic activity is required for the maintenance of genomic stability. (B) Representative images of DAPI staining of the indicated cell lines. Arrows indicate micronuclei or chromosome bridges. (C) Quantification of the abnormal cells based on DAPI staining as in A and B. Error bars indicate SEM from two independent experiments. Total numbers of cells analyzed were as follows for experiment #1/#2,: control, 150/214; RNAi #2, 201/217; KDM2AWT, 254/199; KDM2AD214A, 166/164; and KDM2AN298A, 221/134. (D) Depletion of KDM2A promotes anchorage-independent growth. Soft agar colony formation assay of KDM2A-depleted or control HT1080 cells. The ability of the indicated HT1080 cell line to grow colonies in methylcellulose is shown. Bar graph indicates number of colonies per field 21 d after seeding. Error bars indicate SEM from three independent experiments. (E) Complementation of KDM2A-depleted HT1080 cells with wild-type KDM2A, but not with catalytically inactive mutants, suppresses anchorage-independent growth. Soft agar colony formation assay as in D using the indicated HT1080 cells. Bar graph shows number of colonies per field 21 d after seeding. Error bars indicate SEM from three independent experiments. (F) Loss of KDM2A promotes Ras-induced oncogenic transformation of p19ARF−/− MEFs. (Top) Western analysis of whole-cell extract (WCE) from p19ARF−/− MEFs stably expressing RAS and control vector or two independent RNAi against the untranslated region of KDM2A transcript. (Bottom) Numbers of colonies formed in methylcellulose for each cell line. Error bars indicate SEM from three independent experiments. (G) KDM2AWT, but not the catalytic mutants, suppresses Ras-induced oncogenic transformation of p19ARF−/− MEFs. (Top) Western analysis of whole-cell extract from p19ARF−/− MEFs transduced with Ras and control vector, KDM2AWT, KDM2AD214A, or KDM2AN298A. (Bottom) Soft agar colony formation assay using the indicated cell lines. Bar graph shows number of colonies per field 21 d after seeding. Error bars indicate SEM from three independent experiments.