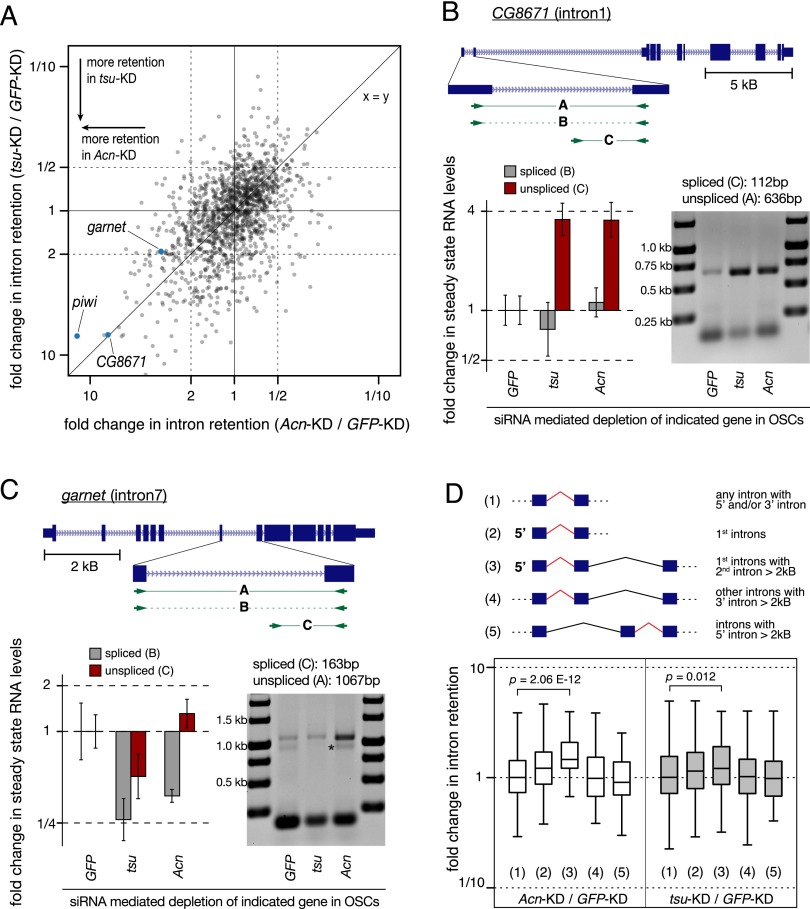
Figure 6.
Transcriptome-wide analysis of EJC-dependent introns. (A) Scatter plot showing the fold changes in the retention of individual introns of OSC transcripts upon Acn or tsu knockdown relative to the control knockdown. Validated hits are indicated (blue dots). piwi int4 is among the most prominent introns that are retained both in Acn and tsu knockdowns. (B,C) Experimental validations of enhanced retention of CG8671 int1 and garnet int7 in tsu- or Acn-depleted OSCs using polyA-selected RNAs. Shown at the top is a cartoon of the respective locus (exons as boxes and introns as lines) with the location of the primers in RT–PCR. The bar charts indicate fold changes in steady-state levels of the indicated CG8671 (B) or garnet (C) transcripts. (Gray) Spliced; (red) unspliced. Amplicon identity is given above. The Agarose/EtBr gel images show RT–PCR products obtained with primers mapping to the flanking exons. The asterisk in C marks a nonspecific product. (D, top) The cartoon depicts the analyzed groups of introns, considering their location in the transcript and the identity of flanking introns. The box plot shows fold changes in intron retention for the groups defined above upon Acn or tsu depletion. First introns followed by a large second intron (group 3) are significantly more retained in EJC-depleted cells (P-values are calculated by the Wilcoxon rank-sum test).