Figure 7. Kir2 channels have a caveolin binding motif at the interface between the TM and cytosolic domains.
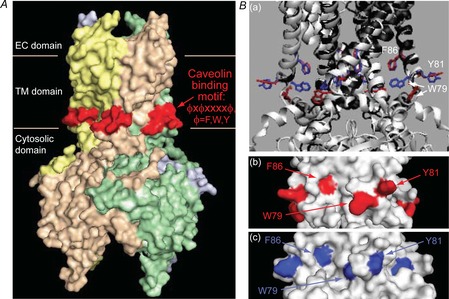
A, location of the caveolin ϕXϕXXXXϕ binding motif (red) at the interface between the outer transmembrane helix and the N-terminus of the channels (residues 81–88 in Kir2.1) in a surface presentation of the crystal structure of Kir2.2. Ba, comparison of the locations of the aromatic residues of the caveolin binding motif in the closed conformation of Kir2.2 (PDB ID 3jyc; the ribbon representation of the channel is coloured black and the aromatic residues in a stick representation are red) and in the conformation obtained in the presence of PIP2 that activates the channel (PDB ID 3spi; the ribbon representation of the channel is coloured white and the aromatic residues in a stick representation are blue). The structures were generated and aligned using the Visual Molecular Dynamics package. b–c surface representations of the region in Kir2.2 that includes the caveolin binding motif showing the locations of the aromatic residues depicted in Fig. 6Ba, in b the closed conformation (red) and Bc the conformation obtained in the presence of PIP2 (blue). Fig. 6A, and 6Bb–c, were generated using Pymol. PIP2, phosphatidylinositol 4,5-bisphosphate.
