Figure 3. A SILAC-based quantitative proteomics experiment to identify putative SUMO-2 substrates and their regulation upon infection by Shigella flexneri.
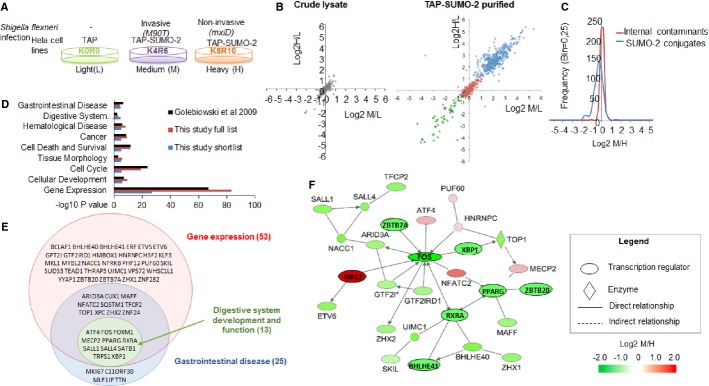
A Overview of the quantitative experiment comparing TAP-expressing with TAP-SUMO-2-expressing HeLa cells that were infected with invasive strain of Shigella (M90T) or infected with non-invasive Shigella mutant (mxiD) for 1 h (n = 1).
B Scatter plots comparing M/L and H/L ratios proteins identified in crude cell lysates (left) and from the TAP-SUMO-2 purified samples (right). In the TAP-SUMO-2 plot, each identified proteins is shown as either external contaminant (in green), internal contaminant (in red) or TAP-SUMO-2 conjugate (in blue).
C Frequency histogram comparing M/H ratios (invasive/non-invasive) between the TAP-SUMO-2 putative substrates and the internal contaminants.
D IPA comparative analysis for functions and diseases associated with the 87 significantly regulated in M/H ratio putative TAP-SUMO-2 conjugates (blue), all 438 TAP-SUMO-2 putative conjugates identified in this study (red) and the 753 putative TAP-SUMO-2 conjugates identified by Golebiowski et al 18 (black).
E Overlap between enriched functional groups identified by IPA for the shortlisted group of 87 TAP-SUMO conjugates. Proteins are shown by gene name.
F Twenty-eight of the 53 transcriptional regulators from the shortlist formed an IPA functional network, implying that the modification state of functionally related proteins involved in gene expression is significantly different between cells invaded by Shigella in comparison with those that are not.
