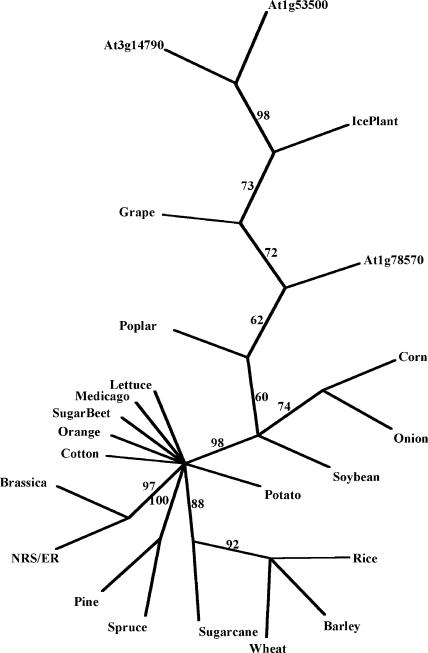
Figure 3.
Phylogenetic analysis of NRS/ER proteins. NRS/ER-like protein sequences, translated from dbEST, were aligned with ClustalX software. Alignments were analyzed with the PAUP software (Swofford, 1998) to generate an unrooted tree. Percentage bootstrap values of 1,000 replicates are given at each branch point. Branch lengths are to scale. Accession numbers are in parentheses: cotton (Gossypium arboreum, BE052407); wheat (Triticum aestivum, CD875945); loblolly pine (Pinus taeda, CF386165); sweet potato (Ipomea batatas, CB330308); soybean (Glycine max, CA784305); orange (Citrus sinensis, CB292447); lettuce (Lactuca sativa, BQ871492); grape (Vitis vinifera, CF512199); barley (Hordeum vulgare, BG369398); medicago (Medicago truncatula, BG587952); brassica (Brassica napus, CD839832); sugarcane (Saccharum officinarum, CA258761); onion (Allium cepa, CF443605); ice plant (Mesembryanthemum crystallinum, BF479490); spruce (Picea mariana, AF051236); sugar beet (Beta vulgaris, BF011071); poplar (Populus tremuloides, CF119362); rice (Oryza sativa, AK071766); corn (Zea mays, AY108467); and the conserved amino acid NRS/ER sequence domain within three Arabidopsis genes annotated At3g14790, At1g53500, and At1g78570.