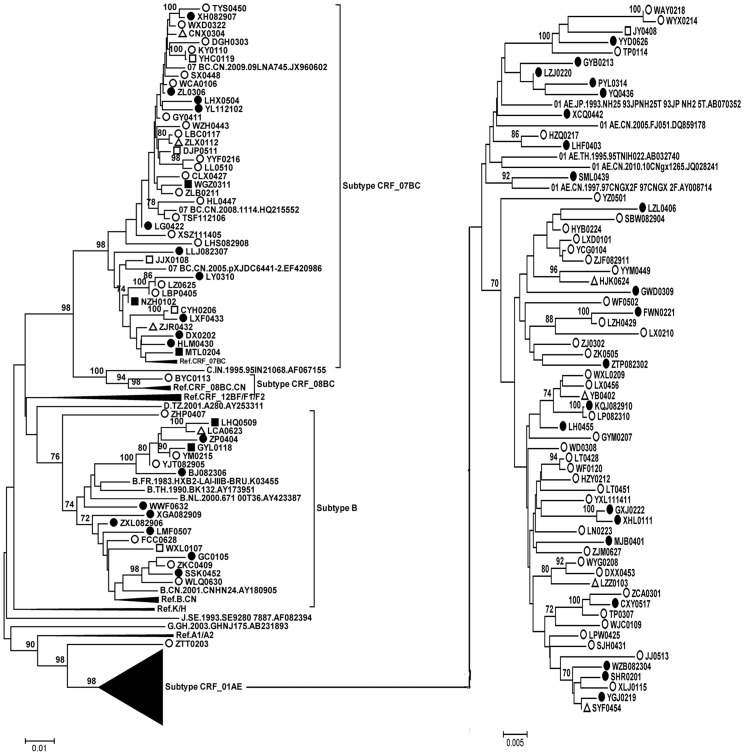
Figure 2. Phylogenetic tree analysis from ART-naïve patients diagnosed between 2009–2011.
122 sequences of HIV-1 gag-pol were used and the lengths were 1057 bp by using HXB2 as the reference genomic. Each reference sequence is labeled with the HIV-1 subtype, followed by country, year, sampling name, and accession number. Isolate ZTT0203 was not clustered with the basic reference subtypes/CRFs. The trees were constructed by the neighbor-joining method. Bootstrap values were calculated from 1,000 analyses, and bootstrap probabilities greater than 70% are indicated at the corresponding nodes of the tree. Open circle: homosexual contact, solid circle: heterosexual contact, open square: blood transmission, solid square: IDU, open triangle: unknown.