Abstract
An intricate history of human dispersal and geographic colonization has strongly affected the distribution of human pathogens. The pig tapeworm Taenia solium occurs throughout the world as the causative agent of cysticercosis, one of the most serious neglected tropical diseases. Discrete genetic lineages of T. solium in Asia and Africa/Latin America are geographically disjunct; only in Madagascar are they sympatric. Linguistic, archaeological and genetic evidence has indicated that the people in Madagascar have mixed ancestry from Island Southeast Asia and East Africa. Hence, anthropogenic introduction of the tapeworm from Southeast Asia and Africa had been postulated. This study shows that the major mitochondrial haplotype of T. solium in Madagascar is closely related to those from the Indian Subcontinent. Parasitological evidence presented here, and human genetics previously reported, support the hypothesis of an Indian influence on Malagasy culture coinciding with periods of early human migration onto the island. We also found evidence of nuclear-mitochondrial discordance in single tapeworms, indicating unexpected cross-fertilization between the two lineages of T. solium. Analyses of genetic and geographic populations of T. solium in Madagascar will shed light on apparently rapid evolution of this organism driven by recent (<2,000 yr) human migrations, following tens of thousands of years of geographic isolation.
Introduction
The pig tapeworm Taenia solium (Cestoda: Taeniidae) is an etiologic agent of cysticercosis, an important zoonosis and neglected tropical disease, and recently ranked as the most important food-borne parasites on a global scale [1]. The lifecycle of T. solium includes humans as the only definitive hosts and domestic pigs as principal intermediate hosts. Cysticercosis refers to infection of various tissues of swine or humans with cysticerci larvae due to ingestion of eggs released from people harboring adult worms in the intestine. Cysticercosis of the central nervous system (neurocysticercosis or NCC), warrants special attention because it is a major cause of seizures and epilepsy in endemic areas [2] and can be lethal especially in remote areas of developing countries [3]. T. solium is distributed worldwide where local people consume pork without meat inspection. We previously reported that T. solium can be divided into two mitochondrial (mtDNA) genetic linages, Asian and Afro-American which differ in the clinical manifestations of human cysticercosis [4]. Their distributions are geographically disjunct in Asia or Africa and Latin America [4]. It has been postulated that T. solium emerged from Africa with early modern humans and through geographic expansion became distributed initially across Eurasia prior to the advent of agriculture and domestication of swine [5]–[7]. Phylogenetic studies have suggested that divergence of the two lineages occurred in the Pleistocene [4], [6], [8]. Recently, sympatry of both mitochondrial lineages was confirmed in Madagascar [6], [8].
Madagascar is a country known to be hyper-endemic for cysticercosis [9], [10]. Cysticercosis in pigs results in condemnation of carcasses, particularly in heavy infections, and thus constitutes a considerable economic challenge. Understanding the current distribution for these parasites and the historical factors involved in geographic colonization of Madagascar can contribute insights of importance in developing a capacity for control and mitigation of infections in swine and human hosts.
Malagasy people are divided into 18 ethnic groups and have diverse cultures. Surprisingly, the first human settlement occurred approximately 2000 years ago as one endpoint of Austronesian migration. Linguistic and archeological evidence suggests that the Malagasy people have mixed ancestry from Island Southeast Asia (ISEA), especially Borneo, and from East Africa [11]; dual origins confirmed by analyses of mtDNA and nuclear DNA [12]. In addition, a contribution to the gene pool of Malagasy people from India has recently been suggested by mtDNA genetic analysis [13]. Prehistoric human migrations can also be traced by parasitological evidence. For example, archaeoparasitology of some intestinal parasites have indicated the existence of human migration routes into the New World other than those involving Bering Land Bridge [14]. Phylogenetic analysis suggested that T. solium has been introduced into Madagascar multiple times from a number of different areas [8], but the dynamics of these introductions and establishment were not fully elucidated. In the present study, reciprocal insights for the distributional history of hosts and parasites emerge from an exploration of T. solium and human occupation of Madagascar.
Historically disjunct populations of T. solium are now in sympatry in Madagascar, affording a unique opportunity to explore the possibility of cross-fertilization and hybridization as a fundamental process among cestodes, and concurrently reflect on the degree of isolation and distinct nature of these genotypes. Cestodes are hermaphrodites with two potential modes of reproduction, self- and cross-fertilization. T. solium has often been referred to as a self-fertilizer because it is nearly always found alone in the human intestine. However, random amplified polymorphic DNA showed heterozygosity in cysticerci of T. solium, suggesting cross-fertilization between different individual worms [15]. Consequently, it may be assumed that the two genotypes of T. solium can cross-fertilize in infections involving multiple adults, which may occur early in the infection process. Analysis of maternal inherited mtDNA alone, however, is not sufficient to examine putative hybridization events. Thus, we initially established nuclear DNA markers to differentiate geographic variation in T. solium. Secondarily, genetic polymorphism of T. solium in Madagascar was investigated to clarify whether hybridization occurs on the island.
Materials and Methods
Parasite isolates and DNA sequencing
During 2005 to 2008, 57 pigs slaughtered from 16 different localities in 5 provinces on Madagascar were found positive for T. solium cysticerci. No specific permissions were required for the field survey, and it did not involve endangered or protected species. Meat inspectors in each province were requested to collect infected pig meats at slaughterhouses from the various locations. Pigs were regularly slaughtered at the official slaughterhouses of each city (Table S1), and the slaughtering was controlled by meat inspectors according to the regulations of the Republic of Madagascar. Pigs were sacrificed for routine slaughterhouse purposes and not for research purposes. When positive for Taenia cysticerci, infected meats were cut and inserted into sterile containers, and sent to the Pasteur Institute of Madagascar within 24 hours. Then the cysticerci were extracted and washed at the laboratory, and frozen at −20°C until use. All samples were then fixed with 70% ethanol and shipped to Japan according to the research agreement between Pasteur Institute of Madagascar and Asahikawa Medical University. One or two cysts from each pig were subjected to molecular analysis. The genomic DNA of each cyst was extracted by DNeasy blood and tissue kit (Qiagen), and subsequently used as a template for polymerase chain reaction (PCR). For the mtDNA gene markers, the entire cytochrome c oxidase subunit I (cox1) and cytochrome b (cob) were amplified by PCR using previously reported primer pairs [4]. PCR products were treated with illustra ExoStar (GE Healthcare) to remove excess primers and dNTPs, and directly sequenced with a BigDye Terminator v3.1 and a 3500 DNA sequencer (Life Technologies).
Nuclear gene markers including RNA polymerase II second largest subunit (rpb2), phosphoenolpyruvate carboxykinase (pepck), DNA polymerase delta (pold) and a low-molecular-weight glycoprotein antigen (Ag2) were amplified using primer pairs published previously [16], [17]. These nuclear genes were chosen because they have been shown to be useful for the molecular phylogeny of taeniid tapeworms including species of Taenia (rpb2, pepck and pold) or for differentiating geographic genotypes of T. solium (Ag2). Initially, 41 geographic isolates of T. solium from 14 countries were used to investigate the geographical variability of nuclear gene markers. PCR products were sequenced with the same protocols as mtDNA gene markers. When geographical variations were found, new primers were designed to amplify the short fragments including mutation sites in order to reduce the cost and labor. PCR was performed in 20 µL volumes containing 0.5 units of Ex Taq Hot Start Version (TaKaRa, Japan), 0.2 mM of dNTP, 1×Ex Taq Buffer with a final MgCl2 concentration of 2.0 mM, 15 pmol of each primer and 1.0 µL of genomic DNA. PCR amplification consisted of initial denaturation of 94°C for 2 min, 35 cycles of 94°C for 15 sec, 55°C for 15 sec and 72°C for 30 sec, and a terminal extension at 72°C for 1 min. In cases of double peaks in the sequencing of nuclear genes, PCR products were ligated into pGEM-T plasmid vector (Promega) and then introduced into Escherichia coli DH5α. At least 10 clonal colonies were picked from an agar plate and their insert DNAs were sequenced to confirm allelic polymorphism.
Data analysis
Nucleotide sequences of the mitochondrial cob (1068 sites) and cox1 (1620 sites) were concatenated into a total sequence (2688 sites). They were aligned by Clustal W 2.0 [18] with those sequences available in public databases. Amino acid sequences were inferred with reference to the echinoderm mitochondrial genetic code [19]. Pairwise divergence values among the obtained nucleotide sequences were calculated using the MEGA5 package [20] using Kimura's two parameter model with a γ-shaped parameter (α = 0.5). The identification of mtDNA haplotypes and the drawing of their network was computed by TCS 1.2 software [21] using statistical parsimony [22]. Evaluation of the rate of outcrossing was based on an estimate of the inbreeding coefficient for each nuclear locus and deviation from Hardy-Weinberg proportions as F = 1-Hobs/Hexp, where H is the actual population heterozygosity and Hexp is the expected heterozygosity under H – W equilibrium.
Results
Mitochondrial DNA phylogeography
In the present study, we collected 109 cysticerci larvae from 57 pigs across 5 provinces on Madagascar. In total, 8 haplotypes (MDG1 to MDG8) of concatenated cox1 and cob genes were detected. When compared with individual genes, the numbers of haplotypes were reduced to 3 (cob) and 7 (cox1). All the nucleotide sequences of each haplotype are deposited in GenBank with accession numbers AB781355-AB781364. The frequency of the nucleotide substitution was 1.6% (17 sites/1068 sites) in cob and 1.4% (22/1620) in cox1 (Tables S2 and S3). Among 39 point mutation sites identified, 24 (61.5%) were synonymous and 15 (38.5%) were non-synonymous substitutions. The maximum value of divergence among the 8 haplotypes was 1.4%. Among the mtDNA gene sequences of T. solium deposited in the public databases, 14 sets of the complete cob and cox1 gene sequences were concatenated and used for the haplotype network analysis together with those from Madagascar (Table 1). These sequences were chosen because they had unequivocal published references allowing confirmation that the sequences of the two genes were obtained from one individual parasite.
Table 1. Mitochondrial haplotypes of T. solium used for the phylogeographic analysis.
| Haplotypesa | Localities | Accession numbers | References | |
| Cox1 | Cob | |||
| MDG1 | Madagascar | AB781355 | AB781362 | This study |
| MDG2 | Madagascar | AB781356 | Same as MDG1 | This study |
| MDG3 | Madagascar | Same as MDG1 | AB781363 | This study |
| MDG4 | Madagascar | AB781357 | Same as MDG1 | This study |
| MDG5 | Madagascar | AB781358 | Same as MDG1 | This study |
| MDG6 | Madagascar | AB781359 | Same as MDG1 | This study |
| MDG7 | Madagascar | AB781360 | AB781364 | This study |
| MDG8 | Madagascar | AB781361 | Same as MDG7 | This study |
| CHN1 | China | AB066485 | AB066570 | Nakao et al. 20024 |
| CHN2 | China | AB066486 | AB066571 | Nakao et al. 20024 |
| ID-BA | Bali, Indonesia | AB631045 | Not determined | Swastika et al. 201224 |
| ID-PA | Papua, Indonesia | AB066488 | AB066573 | Nakao et al. 20024 |
| IND | India | AB066489 | AB066574 | Nakao et al. 20024 |
| NPL1 | Nepal | AB491985 | AB781746 | Yanagida et al. 201023 |
| NPL2 | Nepal | AB491986 | Same as MDG1 | Yanagida et al. 201023 |
| THA | Thailand | AB066487 | AB066572 | Nakao et al. 20024 |
| BRA | Brazil | AB066492 | AB066577 | Nakao et al. 20024 |
| CMR | Cameroon | Same as MEX1 | AB066579 | Nakao et al. 20024 |
| ECU | Ecuador | AB066491 | AB066576 | Nakao et al. 20024 |
| MEX1 | Mexico | AB066490 | AB066575 | Nakao et al. 20024 |
| MEX2 | Mexico | FN995657 | FN995661 | Michelet & Dauga 20126 |
| MEX3 | Mexico | FH995658 | FN995662 | Michelet & Dauga 20126 |
| TZA | Tanzania | AB066493 | AB066578 | Nakao et al. 20024 |
The mitochondrial haplotypes were determined based on the concatenated nucleotide sequences of complete cox1 (1620 bp) and cob (1068 bp), except for ID-BA.
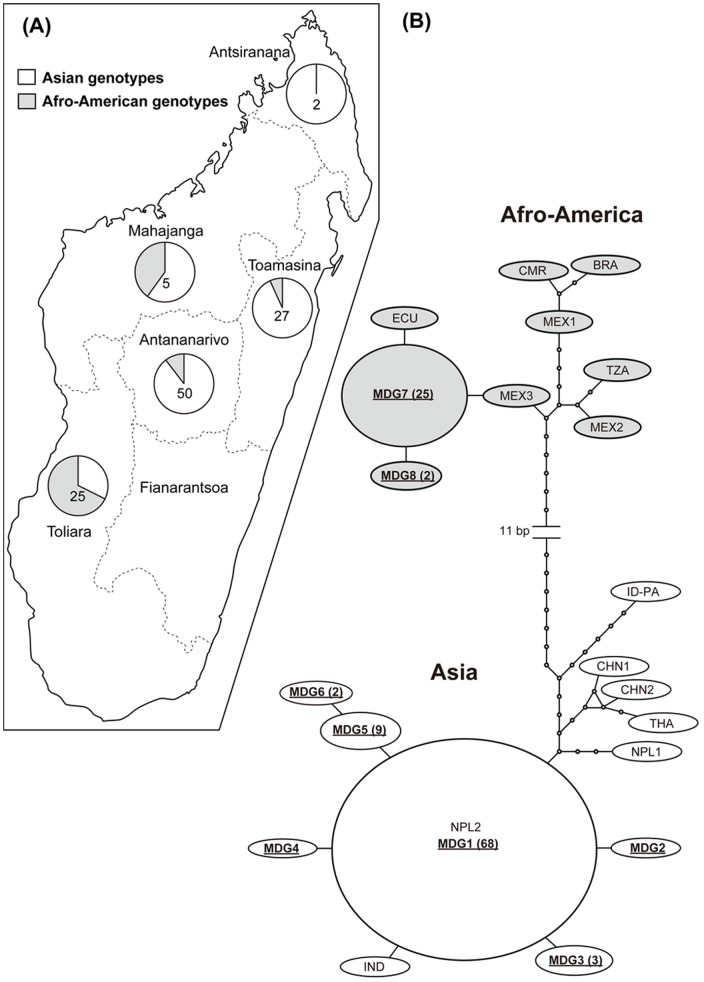
Network analysis clearly showed these 8 haplotypes are divided into two genotypes (Fig. 1). Six haplotypes (MDG1-6) were the Asian genotype and the remaining two (MDG7-8) were the Afro-American genotype. Overall, 77% (84/109) of the Madagascan haplotypes were the Asian genotype (Table 2). The Asian genotype was found in all examined provinces and was generally dominant except in Toliara. The Afro-American genotype was identified in 4 of 5 examined localities. Among the haplotypes obtained, MDG1 was the major (62%), followed by MDG7 (23%). Among 52 pigs in which two cysts were examined, the different haplotypes were simultaneously obtained in 3 hosts; Asian and Afro-American haplotypes (MDG1 and MDG7) were identified from two hosts, and the different Asian haplotypes (MDG1 and MDG4) were obtained from one host. MDG1 was 100% identical to the haplotype obtained from a pig in Nepal [23], and one base different from the Indian haplotype. All Asian haplotypes from Madagascar are grouped with those from the Indian Subcontinent. On the other hand, these Asian haplotypes were distantly related to that from Papua, Indonesia. Further, the cox1 haplotype of the isolate from Bali Island, Indonesia [23] was also distantly related. In contrast, MDG7 was one base different from MDG8 and the haplotypes from Mexico, Ecuador, Bolivia.
Figure 1. Mitochondrial genotypes of T. solium in Madagascar.
(A) Pie charts illustrating the frequencies of the Asian and Afro-American mitochondrial genotypes of T. solium in each collection site. The numbers in the charts show the sample size for parasite isolates examined. Madagascar is divided into the 7 former provinces. (B) The haplotype network of concatenated mtDNA gene sequences. The size of the ellipses is roughly proportional to the haplotype frequency, and the actual numbers of haplotypes (>1) are enclosed in parentheses.
Table 2. Genotypes of T. solium in Madagascar at mitochondrial DNA and each nuclear locus.
| Provinces | No. of pigs | No. of cysts | No. of mtDNA haplotypes | No. of genotypes at each nuclear locus | |||||||||||||||
| Asian | Afro-American | Ag2 | rpb2 | pold | |||||||||||||||
| MDG1 | MDG2 | MDG3 | MDG4 | MDG5 | MDG6 | MDG7 | MDG8 | A/A | A/B | B/B | A/A | A/B | B/B | A/A | A/C | C/C | |||
| Antananarivo | 26 | 50 | 44 | 0 | 0 | 1 | 0 | 0 | 5 | 0 | 46 | 1 | 3 | 49 | 1 | 0 | 47 | 2 | 1 |
| Toamasina | 14 | 27 | 13 | 0 | 1 | 0 | 9 | 2 | 2 | 0 | 27 | 0 | 0 | 27 | 0 | 0 | 27 | 0 | 0 |
| Mahajanga | 3 | 5 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 5 | 0 | 0 | 4 | 1 | 0 | 4 | 1 | 0 |
| Toliara | 13 | 25 | 8 | 1 | 0 | 0 | 0 | 0 | 14 | 2 | 6 | 3 | 16 | 6 | 2 | 17 | 7 | 5 | 13 |
| Antsiranana | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 |
| Total | 57 | 109 | 68 | 1 | 3 | 1 | 9 | 2 | 23 | 2 | 86 | 4 | 19 | 88 | 4 | 17 | 87 | 8 | 14 |
Nuclear DNA
Among the Ag2, rpb2 and pold locus, two (Ag2 and rpb2) or three (pold) alleles were confirmed from the 14 geographical isolates from 14 countries; no geographical variation was found in the pepck locus. Subsequently, Ag2, rpb2 and pold were chosen as appropriate nuclear DNA markers to discriminate the Asian and Afro-American genotypes of T. solium. To amplify the target regions including the variable sites, new primers were designed for rpb2 and pold (Table S4). An additional 27 geographical isolates were analyzed using these new primer sets, to confirm geographical variation. Ag2A, rpb2A and poldA (Asian alleles) were only found in Asia and Ag2B, rpb2B, poldB and poldC (Afro-American Alleles) were obtained from Latin American and African countries (Table 3). The sequence difference among the alleles was 1–3 bp. All the nucleotide sequences of each allele of rpb2 and pold are deposited in GenBank with accession numbers AB781365-AB781369.
Table 3. Distribution of alleles at each nuclear locus around the world.
| Localities | No. isolates examined | Alleles | ||
| Ag2 | rpb2 | pold | ||
| China | 4 | Ag2A | rpb2A | poldA |
| Thailand | 2 | Ag2A | rpb2A | poldA |
| Papua, Indonesia | 2 | Ag2A | rpb2A | poldA |
| Nepal | 3 | Ag2A | rpb2A | poldA |
| India | 4 | Ag2A | rpb2A | poldA |
| Vietnam | 1 | Ag2A | rpb2A | poldA |
| Asian total | 16 | |||
| Tanzania | 7 | Ag2B | rpb2B | poldB |
| Mozambique | 7 | Ag2B | rpb2B | poldB |
| South Africa | 2 | Ag2B | rpb2B | poldC |
| Cameroon | 4 | Ag2B | rpb2B | poldC |
| Mexico | 1 | Ag2B | rpb2B | poldB |
| Ecuador | 2 | Ag2B | rpb2B | poldC |
| Peru | 1 | Ag2B | rpb2B | poldC |
| Brazil | 1 | Ag2B | rpb2B | poldC |
| Afro-American total | 25 | |||
Establishment of nuclear DNA markers allowed us to investigate possible hybridization events in Madagascar. All three nuclear genes were amplified and sequenced for the same 109 cysts as mtDNA genes. Overall, the Asian alleles were the majority in Madagascar with frequencies of 0.81–0.84 (Table 2). Asian alleles were the majority in all the examined regions except for Toliara, and the frequencies of Afro-American alleles in the region were 0.60–0.72. No new alleles were identified among these three loci. Among 12 cysts, the nucleotide sequences of one or more loci could not be determined by direct sequencing because of double peaks in the sequence electropherograms. As the result of cloning of the polymorphic PCR amplicons, two alleles were detected at an approximate ratio of 1∶1. These cases were considered to be heterozygous in each locus. Two cysts obtained from one pig were heterozygous at the all three loci examined. Twenty-two cysts possessed discordant mitochondrial and nuclear genotypes, Asian and Afro-American, in at least one nuclear locus (Table 4). The inbreeding coefficient (F) was estimated only for the sub-population in Toliara because of the considerably biased allele frequency in the other sub-populations; at this locality, F was equal to 0.86 (Ag2), 0.90 (rpb2) and 0.79 (pold).
Table 4. Genotypes of T. solium showing nuclear-mitochondrial discordance.
| ID of samples | MtDNA haplotypea | Genotype at each locusa , b | Localities | ||
| Ag2 | rpb2 | pold | |||
| TsolMDG21b | MDG1 | B / B | B / B | A/A | Toliara |
| TsolMDG29a | MDG1 | B / B | A/A | A/A | Toamasina |
| TsolMDG62a | MDG1 | A/A | A/A | C / C | Antananarivo |
| TsolMDG62b | MDG1 | A/A | A/A | A /C | Antananarivo |
| TsolMDG67a | MDG1 | B / B | B / B | A /C | Toliara |
| TsolMDG68b | MDG1 | B / B | B / B | C / C | Toliara |
| TsolMDG04a | MDG7 | B / B | A /B | A/A | Antananarivo |
| TsolMDG04b | MDG7 | B / B | A/A | A/A | Antananarivo |
| TsolMDG12b | MDG7 | A/A | A/A | A /C | Antananarivo |
| TsolMDG13a | MDG7 | A /B | A/A | A/A | Antananarivo |
| TsolMDG13b | MDG7 | B / B | A/A | A/A | Antananarivo |
| TsolMDG25a | MDG7 | A /B | A /B | A /C | Toliara |
| TsolMDG25b | MDG7 | A /B | A /B | A /C | Toliara |
| TsolMDG28a | MDG7 | A/A | B / B | C / C | Toliara |
| TsolMDG37a | MDG7 | A/A | A/A | A/A | Toamasina |
| TsolMDG37b | MDG7 | A/A | A/A | A/A | Toamasina |
| TsolMDG50a | MDG7 | A/A | A /B | A/A | Mahajanga |
| TsolMDG50b | MDG7 | A/A | A/A | A /C | Mahajanga |
| TsolMDG21a | MDG7 | B / B | B / B | A /C | Toliara |
| TsolMDG68a | MDG7 | B / B | B / B | A /C | Toliara |
| TsolMDG69a | MDG7 | A /B | B / B | C / C | Toliara |
| TsolMDG69b | MDG7 | B / B | B / B | A /C | Toliara |
Haplotypes and alleles in bold are Afro-American ones.
Genotypes with underline indicate those at heterozygous loci.
Discussion
The sympatric distribution of Asian and Afro-American mitochondrial genotypes was confirmed on Madagascar, corroborating a prior report [6], [8]. Although the Afro-American mitochondrial genotype previously was identified only in Toliara [8], we confirmed the co-occurrence of Asian and Afro-American genotypes in 4 out of 7 provinces, indicating a widespread distribution for the two mitochondrial genotypes across the island. Major genotypes differed geographically and across provinces. The Asian genotype was generally dominant at all localities except in Toliara, where 64% of the parasite isolates were the Afro-American genotype.
Differences in the distribution of the dominant genotypes of T. solium among provinces can be attributed to disparate history and ethnic origins in each region and patterns of human dispersal and migration over the past several thousand years. Phylogenetic analyses of Taenia have suggested a relatively deep origin in Africa for T. solium, which may have initially parasitized hominin ancestors of modern humans in the early Pleistocene following a host-switching event from large carnivores [5], [7], [25]. It has been postulated that T. solium emerged from Africa with early modern humans and through geographic expansion became distributed initially across Eurasia prior to the domestication of swine which now represent a primary intermediate host [5], [6]. Although there is no direct evidence, phylogenetic studies using mtDNA markers have suggested the divergence of the two genotypes, now associated respectively with Africa/America and with southern Asia/Indian Subcontinent occurred in the Pleistocene [4], [6], [8].
The dominant haplotype in Madagascar (MDG1) demonstrates Asian affinities and is genetically most similar to those from Nepal and India, but distantly related to that from Papua, Indonesia. Further, a cox1 gene sequence of the isolate from Bali Island [24] was distantly related to MDG1 and other haplotypes from Madagascar. Consequently, it appears that the origin of the Asian genotype on Madagascar is not from ISEA, coincidental with the first human immigrants, but from the Indian Subcontinent. Although Asian origins of the Malagasy people have generally been linked to immigrants and populations from ISEA, our result and recent report on human mitochondrial genetics [13] indicate the importance of Indian influence on the diversity of people and culture in Madagascar consistent with and reflecting a history of human dispersal within the past 2,000 years.
On the other hand, the dominant Afro-American haplotype in Madagascar (MDG7) is closely related to those from Mexico and Ecuador. It does not imply a direct link for Madagascan and Latin American populations, because it is apparent that Afro-American haplotypes have been widely disseminated and the same haplotype can be obtained from both African and Latin American countries [4], [8]. It was suggested that T. solium was introduced into Latin America from Europe or Africa coincidental with European expansion and development of maritime trade routes after the 15th century [4], [26]. The dominance of the Afro-American genotype at Toliara, where the current populace is primarily of African descent, suggests that parasites were introduced to Madagascar, probably recurrently, with people and swine from coastal East Africa in a time frame within the past hundreds of years, although clarification requires further study of isolated populations in areas bordering the Mozambique Channel.
Both Asian and Afro-American genotypes on Madagascar showed a simple network with the major (MDG1 and MDG7) and satellite haplotypes. This result indicates a minimum of two independent events of anthropogenic introduction for T. solium from historically disjunct geographic regions in relatively shallow ecological time. It is not clear whether T. solium was introduced with infected pigs or humans, but it is reasonable to consider that establishment occurred after the first human settlement 2000 years ago because humans are the only definitive hosts. Phylogeography of swine has revealed the distribution of different haplogroups among South Asia, mainland Southeast Asia and ISEA, resulting from Neolithic, human-mediated translocation [27], [28]. Thus, genetic analysis of the pigs in Madagascar may shed light on how the tapeworm dispersed across the Indian Ocean.
In the present study, nuclear-mitochondrial discordance was confirmed in all three loci examined, suggesting hybridization between individual worms possessing different genotypes in the recent past. Two cysts from a pig in Toliara were heterozygous at all three loci, suggesting these were F1 hybrids between Asian and Afro-American populations; this genotype could appear at the F2 or later generation by self-fertilization of a hybrid-derived individual worm. Nuclear-mitochondrial discordance in T. solium has been confirmed only in Madagascar to date, indicating the hybridization event occurred on the island. The inbreeding coefficient F of the sub-population in Toliara was about 0.8–0.9. If F is interpreted as the rate of selfing [29], it means that 10–20% of the parasite individuals in the subpopulation are outcrossing. The frequency of outcrossing is much less than that demonstrated in another taeniid tapeworm Echinococcus granulosus, which were estimated as 74% [30]. Such a contrast is consistent with extraordinarily large infrapopulations typical of E. granulosus in canid definitive hosts and thus the chance of mating is simply higher than that of T. solium. Nevertheless, the estimated rate of outcrossing for T. solium was unexpectedly high when considering that these tapeworms are nearly always found in single-worm infections in humans. However, we experienced a case of taeniasis involving 20 T. solium adults in China [31], and we assume that the multiple infection of T. solium tapeworms is not so rare in endemic areas. Our result suggests that the chance of outcrossing has been underestimated and establishes hybridization as a common outcome for the Asian and Afro-American genotypes in zones of contact or sympatry. Further epidemiological study on taeniasis in Madagascar may contribute to a better understanding of the breeding systems of T. solium.
Conclusions
In the present study, we show that T. solium was introduced and established on Madagascar at least twice in the past 2000 years. An Asian origin, from the Indian Subcontinent, for some genotypes of T. solium contrasts with the established history and ancestry of the Malagasy culture primarily from ISEA. Our results demonstrate that tapeworms from geographically disjunct regions in Africa or Latin America and the Indian Subcontinent are now in secondary contact on Madagascar following a history of isolation for populations that may extend to the Pleistocene. Parasites with origins in Africa/Latin America or Asia reflect the complex history of development of the Malagasy culture, and in this case provide compelling evidence for the history of human occupation of the island. Our study highlights the importance of elucidating the determinants for distributions of human pathogens and is especially relevant given manifestation of distinct disease syndromes and socioeconomic impact associated with the two recognized genotypes of T. solium [4], [32].
Supporting Information
Location of the slaughterhouses and the numbers of pigs and cysts examined in each location.
(DOC)
Nucleotide substitutions of mitochondrial cob gene in 22 haplotypes of T. solium .
(DOC)
Nucleotide substitutions of mitochondrial cox1 gene in 23 haplotypes of T. solium .
(DOC)
PCR primer pairs used for the amplification of nuclear gene markers.
(DOC)
Acknowledgments
The authors are grateful to Ms. Toshiko Miura and Tomoe Nakayama for their kind support in molecular analyses.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All nucleotide sequence files are available from GenBank (accession numbers AB781355-AB781369).
Funding Statement
This study was supported by the Institut (http://www.pasteur.mg/) Pasteur de Madagascar and by the Japan Society for Promotion of Science (JSPS: http://www.jsps.go.jp/) Asia/Africa Scientific platform (2006-2011), the Grant-in-Aid for Scientific Research from JSPS (21256003, 24256002) and the Special Coordination Fund for Promoting Science and Technology from the Ministry of Education, Japan (2010-2012) to A. Ito. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Robertson LJ, van der Giessen WB, Batz MB, Kojima M, Cahill S (2013) Have foodborne parasites finally become a global concern? Trends Parasitol 29: 101–103. [DOI] [PubMed] [Google Scholar]
- 2. Ndimubanzi PC, Carabin H, Budke CM, Nguyen H, Qian Y-J, et al. (2010) A systematic review of the frequency of neurocyticercosis with a focus on people with epilepsy. PLoS Negl Trop Dis 4: e870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ito A, Nakao M, Wandra T (2003) Human taeniasis and cysticercosis in Asia. Lancet 362: 1918–1920. [DOI] [PubMed] [Google Scholar]
- 4. Nakao M, Okamoto M, Sako Y, Yamasaki H, Nakaya K, et al. (2002) A phylogenetic hypothesis for the distribution of two genotypes of the pig tapeworm Taenia solium worldwide. Parasitology 124: 657–662. [DOI] [PubMed] [Google Scholar]
- 5. Hoberg E (2006) Phylogeny of Taenia: species definitions and origins of human parasites. Parasitol Int 55: S23–S30. [DOI] [PubMed] [Google Scholar]
- 6. Michelet L, Dauga C (2012) Molecular evidence of host influences on the evolution and spread of human tapeworms. Biol Rev 87: 731–741. [DOI] [PubMed] [Google Scholar]
- 7. Terefe Y, Hailemariam Z, Menkir S, Nakao M, Lavikainen A, et al. (2014) Phylogenetic characterisation of Taenia tapeworms in spotted hyenas and reconsideration of the “Out of Africa” hypothesis of Taenia in humans. Int J Parasitol 44: 533–541. [DOI] [PubMed] [Google Scholar]
- 8. Michelet L, Carod JF, Rakontondrazaka M, Ma L, Gay F, et al. (2010) The pig tapeworm Taenia solium, the cause of cysticercosis: Biogeographic (temporal and spacial) origins in Madagascar. Mol Phylogenet Evol 55: 744–750. [DOI] [PubMed] [Google Scholar]
- 9. Mafojane NA, Appleton CC, Krecek RC, Michael LM, Willingham AL III (2003) The current status of neurocysticercosis in Eastern and Southern Africa. Acta Trop 87: 25–33. [DOI] [PubMed] [Google Scholar]
- 10. Rasamoelina-Andriamanivo H, Porphyre V, Jambou R (2013) Control of cysticercosis in Madagascar: Beware of the pitfalls. Trends Parasitol 29: 538–547. [DOI] [PubMed] [Google Scholar]
- 11. Dewar RE, Wright HT (1993) The culture history of Madagascar. J World Prehist 7: 417–466. [Google Scholar]
- 12. Hurles M, Sykes B, Jobling M, Foster P (2005) The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am J Hum Genet 76: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dubut V, Cartault F, Payet C, Thionville M, Murail P (2010) Complete mitochondrial sequences for haplogroups M23 and M46: insights into the Asian ancestry of the Malagasy population. Hum Biol 81: 495–500. [DOI] [PubMed] [Google Scholar]
- 14. Araujo A, Reinhard KJ, Ferreira LF, Gardner SL (2008) Parasites as probes for prehistoric human migrations? Trends Parasitol 24: 112–115. [DOI] [PubMed] [Google Scholar]
- 15. Maravilla P, Gonzalez-Guzman R, Zuniga G, Peniche A, Dominguez-Alpizar JL, et al. (2008) Genetic polymorphism in Taenia solium cysticerci recovered from experimental infections in pigs. Infect Genet Evol 8: 213–216. [DOI] [PubMed] [Google Scholar]
- 16. Knapp J, Nakao M, Yanagida T, Okamoto M, Saarma U, et al. (2011) Phylogenetic relationships within Echinococcus and Taenia tapeworms (Cestoda: Taeniidae): An inference from nuclear protein-coding genes. Mol Phylogenet Evol 61: 628–638. [DOI] [PubMed] [Google Scholar]
- 17. Sato MO, Sako Y, Nakao M, Wandra T, Nakaya K, et al. (2011) A possible nuclear DNA marker to differentiate the two geographic genotypes of Taenia solium tapeworms. Parasitol Int 60: 108–110. [DOI] [PubMed] [Google Scholar]
- 18. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 19. Nakao M, Sako Y, Yokoyama N, Fukunaga M, Ito A (2000) Mitochondrial genetic code in cestodes. Mol Biochem Parasit 111: 415–424. [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- 22. Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yanagida T, Yuzawa I, Joshi DD, Sako Y, Nakao M, et al. (2010) Neurocysticercosis: assessing where the infection was acquired from. J Trav Med 17: 206–208. [DOI] [PubMed] [Google Scholar]
- 24. Swastika K, Dewiyani CI, Yanagida T, Sako Y, Sudarmaja M, et al. (2012) An ocular cysticercosis in Bali, Indonesia caused by Taenia solium Asian genotype. Parasitol Int 61: 378–380. [DOI] [PubMed] [Google Scholar]
- 25. Hoberg E, Alkire N, Queiroz A, Jones A (2001) Out of Africa: origins of the Taenia tapeworms in humans. P Roy Soc Lond B Bio 268: 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez-Hernandez F, Jimenez-Gonzalez D, Chenillo P, Alonso-Fernandez C, Maravilla, et al (2009) Geographical widespread of two lineages of Taenia solium due to human migrations: Can population genetic analysis strengthen this hypothesis? Infect Genet Evol 9: 1108–1114. [DOI] [PubMed] [Google Scholar]
- 27. Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, et al. (2005) Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307: 1618–1621. [DOI] [PubMed] [Google Scholar]
- 28. Larson G, Cucchi T, Fujita M, Matisoo-Smith E, Robins J, et al. (2007) Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc Natl Acad Sci USA 104: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCauley DE, Whittier DP, Reilly LM (1985) Inbreeding and the rate of self-fertilization in a grape fern, Botrychium dissectum . Am J Bot 72: 1978–1981. [Google Scholar]
- 30. Haag KL, Marin PB, Graichen DAS, De La Rue ML (2011) Reappraising the theme of breeding systems in Echinococcus: is outcrossing a rare phenomenon? Parasitology 138: 298–302. [DOI] [PubMed] [Google Scholar]
- 31. Ito A, Li T, Chen X, Long C, Yanagida T, et al. (2013) Mini review on chemotherapy of taeniasis and cysticercosis due to Taenia solium in Asia, and a case report with 20 tapeworms in China. Trop Biomed 30: 164–173. [PubMed] [Google Scholar]
- 32. Campbell G, Garcia H, Nakao M, Ito A (2006) Genetic variation in Taenia solium . Parasitol Int 55: S121–S126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of the slaughterhouses and the numbers of pigs and cysts examined in each location.
(DOC)
Nucleotide substitutions of mitochondrial cob gene in 22 haplotypes of T. solium .
(DOC)
Nucleotide substitutions of mitochondrial cox1 gene in 23 haplotypes of T. solium .
(DOC)
PCR primer pairs used for the amplification of nuclear gene markers.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All nucleotide sequence files are available from GenBank (accession numbers AB781355-AB781369).