Abstract
Over the past decade we have seen a growth in the provision of chemistry data and cheminformatics tools as either free websites or software as a service (SaaS) commercial offerings. These have transformed how we find molecule-related data and use such tools in our research. There have also been efforts to improve collaboration between researchers either openly or through secure transactions using commercial tools. A major challenge in the future will be how such databases and software approaches handle larger amounts of data as it accumulates from high throughput screening and enables the user to draw insights, enable predictions and move projects forward. We now discuss how information from some drug discovery datasets can be made more accessible and how privacy of data should not overwhelm the desire to share it at an appropriate time with collaborators. We also discuss additional software tools that could be made available and provide our thoughts on the future of predictive drug discovery in this age of big data. We use some examples from our own research on neglected diseases, collaborations, mobile apps and algorithm development to illustrate these ideas.
Keywords: Cloud, Collaboration, Cheminformatics, Drug Discovery, Mobile Apps
Introduction
A lot can happen in a decade. We have gone from having few if any free resources such as databases of small molecules or software for drug discovery on the web, to literally thousands [1]. For example databases like ChemSpider [2, 3] have grown to house not just tens of millions of molecules but have become repositories for reactions and a vast treasure trove of chemistry data. At the other extreme, commercial offerings running software as a service (SaaS) ventures is nothing new, and being on a vendor-hosted cloud or an internal data center is not new either, even though companies continue to define their products by referring to it. What the pioneers of this approach do next will redefine the types of products we see created for drug discovery for years to come. As an example from our own experiences GeneGo (Thomson Reuters) was one of the earliest providers of drug discovery technologies (MetaDrug, MetaCore [4-10]) related to systems biology and integrated cheminformatics tools as SaaS. Collaborative Drug Discovery, Inc. (CDD) was likely the first to offer a private vault for storing chemistry and biology data as a multitenant SaaS [11]. Meanwhile, other larger software companies have acquired similar smaller companies [12] to give them a presence ‘on the cloud’ and a collaborative software offering. Companies like CDD have built a viable business around the software and grants focused on using their technologies alongside other tools to advance research on neglected and other diseases [11, 13, 14].
Research collaborations are increasingly seen as being key to accelerating biomedical research and these are likely to be facilitated by computational methods [15]. However, we have suggested earlier that scientists are rarely collaborative or open with their data until publication or patenting, due to intellectual property (IP) concerns [13, 16]. Emerging collaborative software technologies allow researchers to specifically draw the line between pre-competitive and competitive areas and data more than previously possible. Scientific collaborations increasingly are becoming more important for the pharmaceutical industry especially in difficult areas or those suggested to have less commercial viability. The industry has had to adapt by acquiring or partnering to bring in innovation or products [17] as well as outsourcing many aspects of R&D. At some point companies have to share their data whether this is during a collaboration, licensing, due diligence, pre-purchase or post-purchase. Each of these processes has challenges when it comes to sharing molecular structures and associated data representing the IP for the companies or research groups involved. Increasingly such groups are involved in multi-organization collaborations such as public-private partnerships (PPP).
As an example of PPPs, CDD is involved in several collaborations such as More Medicines for Tuberculosis (MM4TB), Bill and Melinda Gates Foundation (BMGF), TB Accelerator and the NIH Blueprint for Neuroscience Research (BPN). In all of these initiatives, molecules and screening data with IP are securely shared between collaborators. Other similar software is likely required and available to share genomic and proteomic data but is outside the scope of this discussion. Currently data and associated molecules are selectively shared as a function of complex negotiations. Often important information is then missing for the other groups involved which could help the global goals of the project. Ideally this could be shared too in a way that did not interfere with other projects, IP or relationships outside the scope of the current project of interest. There is also a growing need for public collaborations through initiatives that require open data [3, 18] though some of these may not truly be open themselves e.g. Open Source Drug Discovery [19]. One European IMI funded PPP initiative European Lead Factory [20] is focused on high throughput screening (analogous to what the NIH has funded at its many screening centers), and another initiative, Elixir is a pan European infrastructure for biological information [21]. What cannot be denied is the growing mountain of data in the public domain and the likely growth in the need for collaboration to move projects rapidly and make sense of the accumulated information.
Bigger data and neglected diseases
A decade ago the amount of HTS data available was just a fraction of what it is now. The arrival of PubChem [22] and the mandate of publishing NIH-funded experimental data into this database, has obviously had a big impact, putting thousands of assays and millions of data points onto the internet. But for such data to be valuable it requires the underlying data be consistent, reliable and well-linked. The data also has to be of high quality as errors in structure can multiply from database to database [23, 24]. Then we can apply or build algorithms to mine the data, find patterns in it and help make well-informed decisions. Can we really call this ‘big data’ though? It is all relative as one scientist's big data is another's small data. Relative to many nonscientific fields, what cheminformatics data lacks in size, it makes up for in terms of inconvenience and difficulty of handling. Perhaps we can just call this biomedical related data “bigger data” compared to what we had access to in the past (e.g. tens to hundreds of compounds for quantitative structure activity relationships).
One area in which we are seeing larger amounts of screening data being useful and more accessible is in neglected disease research. Neglected diseases are a group of biologically unrelated diseases that are grouped together because they disproportionately affect marginalized populations or they lack effective treatments or vaccines, or existing products to treat them are not accessible to the populations affected [25]. While the definition of a neglected disease varies, the category generally includes: tuberculosis (TB), malaria, Chagas disease, African sleeping sickness, schistosomiasis, leishmaniasis and others for which there is a lack of economic incentives or “market” to provide motivation for product development [26-28]. Many of the pathogens involved, whether bacterial, parasitic, or viral, have complex life cycles and diverse approaches for evading the host immune system, rendering the development of new drugs and vaccines all the more challenging. Furthermore, these neglected diseases receive a relatively small amount of research investment ($80M to approximately $500M [29]) from governments and pharmaceutical companies in the developed world when we know it costs over a $1 billion to bring a drug to market [30]. The scientific challenges and limited funding available for neglected disease drug discovery and development highlight the importance of doing as much as possible with the data. These diseases are not seen as commercially viable next to major diseases, so many companies donate patents and fund some limited research efforts and participate in PPPs. Currently available data relevant to neglected disease drug discovery is extremely diffuse, existing in an array of public or private databases (e.g. ChemSpider, PubChem, CDD, ChEMBL). One example is Mycobacterium tuberculosis (Mtb) which is the causative agent of TB that has infected approximately 2 billion people, and continues to kill 1.3 million people annually. We are seeing more companies making increasing quantities of screening data publically accessible, as well as the need to collaborate and share this data as GlaxoSmithKline have made 177 compounds with Mtb activity [31] and 14,000 compounds with antimalarial activity [32] available. Surprisingly, we are still making very slow progress in finding novel therapeutics [33] for TB and the clinical pipeline is limited [34]. Ideally we should be learning from the past efforts in TB drug discovery and yet we do not appear to be doing something that is simple yet effective, learning from the data that already exists [35]. The current predominant method for identifying compounds active against Mtb is to use phenotypic high throughput screening (HTS) [36-39] and the hit rate of these screens tends to be in the low single digits. We can estimate that upwards of 5 million compounds have been screened against Mtb over the last 5-10 years [35]. There are around 1500 Mtb hits of interest from one laboratory alone [38-41]. Leveraging this prior knowledge (by curating the data) to produce validated computational models is an approach that can be taken to improve screening efficiency both in terms of cost and relative hit rates. Machine learning and classification methods have been used in TB drug discovery [42], and have enabled rapid virtual screening of compound libraries for novel chemotypes [43, 44]. The use of cheminformatics for tuberculosis drug discovery has been summarized [45-47] and can be readily implemented early in the process as a means to limit the number of compounds needing to be screened, therefore saving time and money [48-52]. Recent publications in this area have hit rates >20% and focus on favorable compounds with low or no cytotoxicity [51, 52]. More recently, combining datasets to use all 350,000 molecules with in vitro data from a single laboratory for computational models has been attempted. Interestingly our recent data suggests that smaller models with thousands of compounds may perform just as well as these “bigger data” models [53].
Throughout all of this work using the Mtb datasets for over 5 years, we have shown how additional value can be generated from such published data. Similar cheminformatics approaches have also been applied to other diseases [54-57]. Computational methods result in cost savings by eliminating the need for some experiments or testing many hypotheses which would not normally be possible without such models. While there has been considerable screening and identification of hits, a possible bottleneck is the progression of compounds and expansion of structure activity relationships that could result in viable leads. To date we estimate that there are ca. 2000 in vitro Mtb hits that need prioritizing before progressing. The in vitro, in vivo and clinical data for TB do not exist in a single database. Our own efforts to collate mouse in vivo data for modeling took many months and were recently described [35]. We see this lack of data coordination as a major limitation to progress. There is also no centralized organization for project management and minimal collaboration or coordination in the field. This suggests that even though we are drowning in data, actually a bigger challenge is the integration and analysis of it before ultimately being able to use it for predictive models and prospective testing. These observations may also be broadly applicable beyond Mtb, but illustrate what can be achieved with generally much larger datasets than were available in the past.
Collaborative sharing of molecules and data
Do we take the importance of privacy concerns for our data too far or not far enough? Should we think more carefully about what is the “real high value” data and perhaps loosen our belts and share more than we hoard data? Should we just find new ways to share data? For example we have already seen several companies compare their compound libraries to each other e.g. Bayer and Schering [58], Bayer and AstraZeneca [59] or in the case of Pfizer to the literature [60] using fingerprints, physicochemical properties and matching/similar compounds to show minimal overlap. While this is not the same as openly sharing molecules and their proprietary data on assays, many companies are involved in PPPs like those described earlier. What steps could be taken to increase the amount of secure data sharing?
Finding new ways to share relevant chemical information about screening data that leaves structures blinded could open the door for increased collaboration. These methods include better strategies for identifying active molecules from primary screens, which leverages information from fingerprints [61], scaffold groupings [62, 63], economic modeling [64-66], and improved processing of raw data [67-69]. They also include automatic methods of organizing screening data into workflows [70] and a series of approaches for visualizing how biological activity maps to chemical space [71-74]. Secure methods of sharing molecules and data could make outsourcing of chemical analysis possible (without sharing the structure itself). Outsourcing is increasingly important in drug discovery because it reduces the cost of many R&D efforts and enables centralization of expertise [75-77]. As more data is made available through these efforts it is possible unexpected connections and patterns in data can be identified that could have an impact on research. These connections certainly are impossible to predict. They include unexpected signals in screening data that indicate either specific molecules or mechanisms by which to treat human disease, or indications that might relate to adverse effects. Sharing large collections of proprietary assay data, with structures blinded, would enable researchers not part of the original data collection process to potentially improve how we do drug discovery. For example, a recent study used a small dataset published in patents from AstraZeneca, to show how different liquid dispensing methods can severely impact the IC50 data generated in high throughput screening and in turn impact the computational models that are built and decisions based on them [78]. Collaboration across multiple pharmaceutical companies and academia could potentially address this on a much larger and more convincing scale, but it likely awaits the use of secure sharing methods that do not reveal structures.
Nearly a decade ago there were attempts at securely sharing molecule-related structure activity relationship data but these stalled when it was suggested that the proposed encryption methods were all fallible. For example, a 2005 American Chemical Society meeting, co-chaired by Dr. Christopher Lipinski and Dr. Tudor Oprea included a session on securely sharing chemical information to support collaborative development of absorption, distribution, metabolism and excretion (ADME) predictors [79-89]. Swamidass and co-workers recently proposed several approaches to the problem of sharing molecules securely [90] that may overcome the previous failings. First, they propose a totally new, secure method of sharing useful chemical information from small-molecule screens, without revealing the structures of the molecules [90]. The method generates scaffold networks for compounds, enabling sharing of: molecule identifiers with assay data; how molecules in a screen are connected to one another in a screening network; how molecules are grouped together into scaffold groups; how these groups are connected into trees; how these groups are connected into networks; and how molecules are connected together into R-group networks. Statistical analysis using the PubChem data also clearly demonstrated that scaffold networks do not convey enough information to reliably reveal chemical structure [90].
A second proposed approach from the same group uses a new, secure way of measuring the overlap between two private datasets. This method uses an algorithm to construct a private dataset's shareable summary, which is called a “cryptoset” [91]. The overlap between two private datasets can be estimated by comparing their cryptosets. At the same time, it is not possible to determine which specific items are in a private dataset from its cryptoset. Unlike other approaches to this problem [92-94], the item-level security arises from statistical properties of cryptosets rather than the secrecy of the algorithm or computational difficulty, so cryptosets can be shared in public, untrusted environments.
We are aware of at least one company, MedChemica which has successfully developed a business model around technology closely related (but not identical) to what Swamidass and co-workers are proposing above. MedChemica successfully negotiated agreements with three big pharma companies (AstraZeneca, Hoffman La Roche, Genentech) to share anonymized match-pair [95] data for the purpose of improving ADME optimization of lead compounds [96]. MedChemica's partners pay them to provide software to process the structures in internal ADME data into an anonymized form, very similar to the R-group networks described earlier. This anonymized data is then transferred to MedChemica, where it is analyzed, and specific rules to guide ADME optimization are extracted. These rules are then offered back to MedChemica's clients to aid in lead optimization.
Approaches like these for secure data sharing need to be integrated into software tools that are used by scientists to store their data to provide confidence when they do decide to share subsets of their data with different collaborators. This is becoming even more apparent as drug companies reach out increasingly to academics to fill the internal research gaps by externalizing their fundamental chemistry, biology and screening research efforts.
Predictive drug discovery
One of the challenges after high throughput screening is to learn as much as possible about the hits or potential probe compounds being developed. Are they cytotoxic? What liabilities do they have? What off-targets do they have? Could we predict as much as possible about the molecules before we invest more time and efforts in them? This obviously assumes that the computational models for absorption, distribution, metabolism, excretion and toxicity (ADME/Tox) we use for particular properties are predictive and cover enough chemistry space. A major parameter to understand is drug metabolism.
Some of the major issues in drug metabolism include identifying: the enzyme/s involved, the site/s of metabolism, the resulting metabolite/s and the rate of metabolism. Methods for predicting human drug metabolism from in vitro and computational methodologies, and determining relationships between the structure and metabolic activity of molecules are also critically important for understanding potential drug interactions and toxicity. The cytochrome P450 (P450) enzymes are of considerable interest both in terms of metabolism and drug-drug interactions. Computational methodologies can be used for prioritization, and uncovering the relationships between the structure and metabolic activity of novel molecules. A recent approach describes a method called XenoSite [97] for building models that predict CYP-mediated sites of metabolism (SOM) for drug-like molecules with predictive accuracies of 87% on average for nine distinct CYP substrate sets. While this approach focused on phase I metabolism it is possible such approaches could be applied to phase II enzymes also.
Introducing such predictive approaches into software that stores screening data or integrating with such tools may be important for creating a pipeline process. This would enable the likely enzymes involved in metabolism to be predicted for a compound. This may be very important for avoiding specific patient populations that are perhaps poor or extensive metabolizers of a drug which could present problems such as hepatotoxicity or lack of efficacy. Being able to provide information on this level for metabolism and other properties like toxicity [98] in software used for storing and sharing chemistry and biology data is likely to be of value in overall decision making. For example there are already efforts like qsardb.org and ochem.eu which enable public model sharing and development [99, 100]. In addition, websites such as Chembench provide models and tools for modeling to registered users [101]. Our earlier work proposed that open source descriptors and algorithms may be comparable with some commercial software, and that this might facilitate more sharing of computational models [102]. There have also been developments such as QSAR-ML which was developed to enable standards for interoperability of QSAR models [103, 104]. One could imagine that software for secure sharing of models could be carried out similarly to that described earlier for data, such that they can be accessed by selected users. None of the websites for creating or storing QSAR models appear to offer this level of selectivity, and many companies may be wary of accessing them without some idea of security. Vendors that can guarantee that a company's IP will be secure are likely to be more successful in getting big pharmaceutical and biotechnology companies to use and share models in this way. Some advantages of sharing models may be that a collaborator can benefit from models developed with your proprietary data, which in turn benefits your shared goals. Sharing models openly with a community may foster addition of a groups own data to update the models and make them more relevant to internal projects if indeed the data were generated under similar conditions. If you were sharing a model and you wanted to ensure that the user could not identify compounds in the training set, you might disable any features that would measure the distance, similarity etc. to compounds in the training set, or at the very least make these outputs fuzzy. It is likely that more work and discussion on secure computational model sharing and development will happen in future.
Future vision
We have previously suggested some of the needs and opportunities for cheminformatics which we termed the “missing pieces” [105]. A decade ago commercial tools and academic tools were virtually the only choices. In recent years we have seen a greater effort towards open source cheminformatics software [104, 106-108]. Also, a decade ago systems biology was piecing together small biology experiments such as protein-protein interactions to understand the “big picture” [109]. Now the amount of data available in some areas of biology (for many diseases or specific targets) is overwhelming. The challenges are to know where to look for the data you need in the first place. It may be feasible to turn this around and say that databases or data sources should be more proactive about making their data more accessible (or telling you what may be of interest). One way to do this is to use different avenues to create more value from the data.
Recently we have taken the approach we have called “appification”, that is to make a discrete molecule dataset available as a mobile application (app). This has become a common theme in the world of software, but is relatively new to structure-centric chemistry data. To our knowledge this was first achieved with the Green Solvents mobile app which used the American Chemical Society Green Chemistry Institute Pharmaceutical Roundtable Solvent Selection Guide (as a PDF document) [110]. This document lists the 60 solvents by chemical name (and excludes structures) and rates the solvents against safety, health, air, water and waste categories with scores from 1 (few issues) to 10 (most concern) with additional color coding (green, yellow and red). This appification involved curation of the public data and development of a novel interface [111]. The limitations in access and utility of the original document encouraged us to recast the content in a novel manner to greatly enhance visibility and availability to practicing chemists. The data was also used to enable predictions for solvents outside the guide. A similar approach has also been taken with data on 800 molecules with known targets in TB [112, 113] to create the app called TB Mobile. The data originated from a dataset in CDD public [14] but it was felt that the impact could be extended by creating a tool that could be useful for scientists and educators. The resultant app enables the user to view the molecules and known targets alongside other data related to the biology of the target. This represents one relatively simple way to bring cheminformatics and bioinformatics together. We have recently also implemented naïve Bayesian models using our own implementation of open source ECFP_6 descriptors in the app, to enable an alternative approach to target prediction as well as clustering molecules [114].
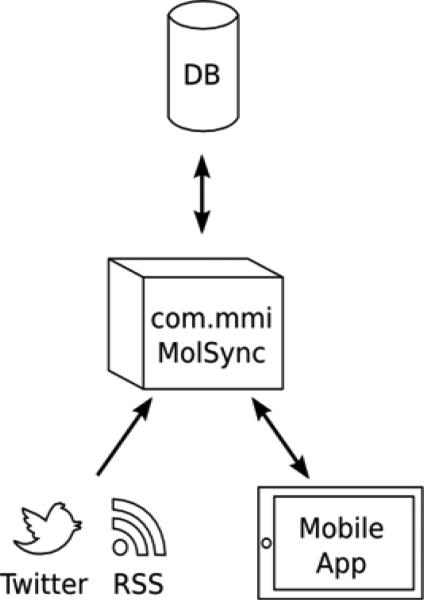
A further novel approach to creating open chemistry and biology databases can be achieved by building on tools we take for granted like Twitter and RSS feeds. A mobile app called Open Drug Discovery Teams (ODDT) [115] harvests Twitter feeds on several hashtags (e.g., #malaria, #tuberculosis, #huntingtons, #hivaids, #greenchemistry, #chagas, #leishmaniasis and #sanfilipposyndrome, and many other additional rare diseases and other topics). Harvesting in this way enables open data and molecules to be collected in an app. You could also think of this as a database with each topic being a subsection (e.g., a database on tuberculosis and a database on malaria etc.). The architecture of the currently deployed ODDT project is shown in Figure 1. The cheminformatics framework that powers the molsync.com web service has been extended to include continual querying of Twitter and RSS feeds for relevant content, and collecting them in a database. We and others have tweeted in to these topics, links to molecules, data and papers. We then added the ability to endorse or reject tweets. In addition, the ability to visualize a fingernail image for each tweet was added, as well as recognition of molecule images and a summary ticker tape.
Figure 1.
ODDT framework
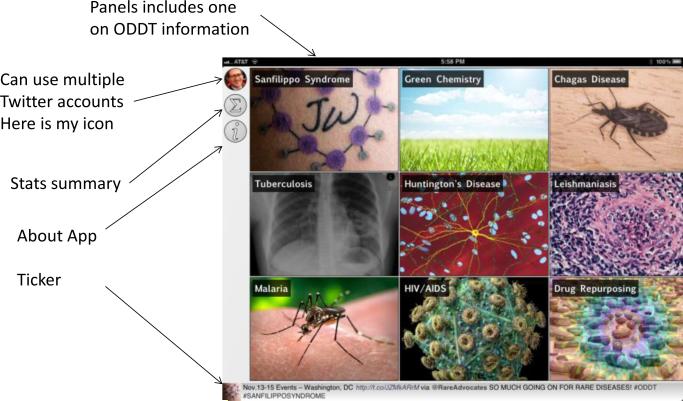
The ODDT app can be used to manage multiple twitter accounts for the user too (Figure 2). The entry screen to the app displays the topics ranked by use. Tapping an image opens a topic on the incoming page and the content is listed on the right. Each tweet can be endorsed and the hyperlinks followed. The “recent” content page in the ODDT app shows entries with at least one endorsement while the content section shows the most popular voted content in rank order. Molecules can be tapped to open in other apps and could be the start of a workflow [116, 117]. If you imagine that one of the hurdles to putting data in public databases is the upload of data files, ODDT represents a simplistic approach enabling true one-click upload of molecules and data via a tweet! Perhaps this is an approach that could be used for secure upload via other messaging systems or direct messaging. It could also be an approach from which the bigger web-based databases could learn.
Figure 2.
Schematic of ODDT mobile app functions.
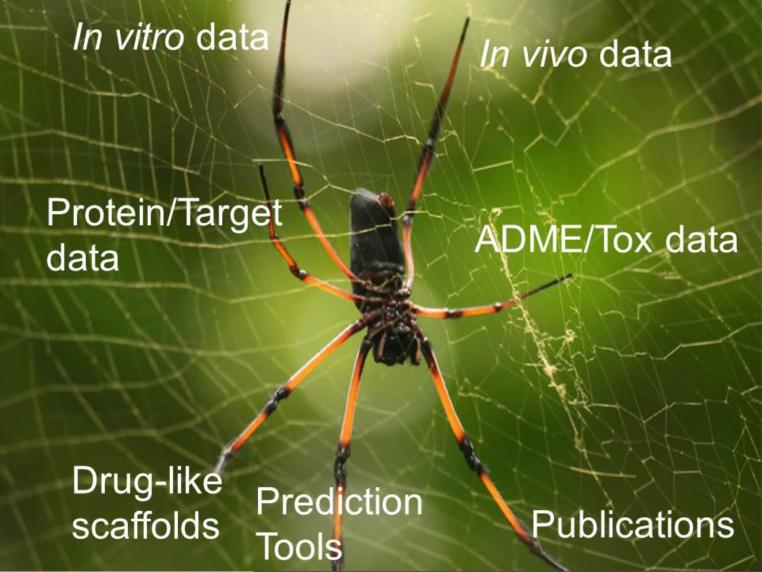
From our experiences in neglected disease research we think there is an opportunity to bring together a range of data and tools (Figure 3) that would facilitate and catalyze the identification of novel therapeutic candidates by combining bioinformatics, cheminformatics data, publications, models and data visualization tools, and curated in vitro and in vivo data. This would enable novel algorithms to be developed to infer candidate drug molecules, targets and mechanisms of drug action. This may in turn allow scientists to generate hypotheses in a single interface. The scientific challenges and limited funding available for neglected disease drug discovery and development highlight the importance of exploring alternative, lower cost approaches to advance drug discovery using cheminformatics, and maximizing the data in the public domain.
Figure 3.
Schematic of the neglected disease related computational tools and information that could be integrated. This could be applicable for any disease or class of diseases.
Other challenges we see as commercial opportunities are how to turn the databases and tools into assistants that make you aware of what data you might want to know about. For example, how can you find collaborators who might have interesting molecules or data? Methods like those described earlier for encrypting or sharing data securely might be valuable in this regard to help you find the data or alert you to its availability. Designing algorithms that can discern the most useful data for connecting researchers could reduce the serendipity involved in building collaborations [118]. Creating a tool that uses social networking features for serious applications such that the software users can “like” a molecule rather than a person might be appealing in some cases for finding researchers with orthogonal preliminary results. Such a system could hasten the pace of research and allow for the sharing of negative data, which is often not published.
Our laboratories (if we still have them) may be like our homes, that is, an “internet of things”. Our databases and software tools should be able to talk wirelessly to devices such as analytical tools and automatically upload data (which we term “no click upload”). Perhaps more likely all of our science will be outside our office. We can leverage contract research organizations (CROs) as well as other contractors via sources like Assay Depot [119] and Science Exchange [120] and our personal connections and networks of collaborators can all do the science we need following our extensive mining of published data [121, 122] and predictions, perhaps even using virtual screening to decide which compounds to test.
How can we use the published data available to help tailor medicines to overcome our own genetic variability and side effects? For example, variability in metabolism is one issue, but what about variability in transporters and regulation of different proteins that can impact drug disposition? We are at a stage where there is increasing interest in computational models for human drug transporters which could be used proactively in the same way that we use models for P450s [123]. Such metabolism and transporter models should probably be used in parallel to profile compounds and predict liabilities and drug-drug and drug-transporter interactions.
Thinking about what is feasible by integrating data on diseases or at least making it available alongside tools to facilitate collaboration and drug discovery, you can begin to think of how non-scientists or non-specialists can leverage them also. For example can we bring non-scientists in to help us develop “outside the box” thinking to tackle tough problems, whether in design of molecules, or biological problems to help cure rare diseases [124, 125]. We need to think about developing new tools that leverage the crowd (Table 1).
Table 1.
Tools for facilitating drug discovery that could leverage the crowd. We may want to consider integrating these multiple features into software tools like Open Drug Discovery Teams [128] that enable access to non-specialists.
| Features | Details |
|---|---|
| Funding research | This enables scientists to post project ideas they want funded. Individuals, foundations (the crowd) could then select and fund this research. Alternatively individuals could post their own ideas for projects they want to see done. The scientists and disease foundations could then engage in dialog. This approach would increase the efficiency of funding research. |
| Crowdsourcing research | Scientists or disease foundations propose work they cannot do and they ask for help. This may be a request for pro bono or paid help. It may be that people with time and flexibility in their careers could simply volunteer their time to a project. |
| Externalizing to companies | This would provide links to CROs and other companies that could assist with various aspects of R&D. |
| Sharing research openly | This could merge efforts like ODDT with a database element, which enables the searching by compounds, by text, storage of molecules etc. It would also bring in open data from external sources from the internet. |
| Precompetitive collaboration | This would be a location that could stimulate such collaborations and provide a location for discussion or to propose projects. Project teams could then self-organize and provide a means for delivering content/projects to be shared. |
| Finding collaborators | This could use tools to enable foundations and parents to search by topic, disease, search grants, and find scientists that can do the research and enable them to connect with them. |
Conclusion
In summary, collaboration and tools to enable data sharing in drug discovery are likely to continue in their importance. Therefore some of the developments we propose in enabling secure or encrypted sharing methods may be important to consider. As databases are integrated or linked together, how we handle and license the data will be key, and some simple rules have already been proposed [126]. The mountain of data available across databases that are either public or private will undoubtedly continue to grow, and this will present challenges we will need to overcome in order to manipulate, mine and model it. We will need some creativity to develop new visualization paradigms that enable insights and lead to the next experiment. On the other hand, as mobile devices continue to expand their utility [127], useful tools and abilities to interact with data are possible, as are extended workflows [117]. While such devices may not be able to handle massive datasets within them just yet, they do present an access point to databases and more powerful tools on the cloud. The utility of being able to take your data with you and explore it on a tablet has some advantages. As we have shown here and elsewhere, mobile devices also represent a way to prototype how we can use published data and cheminformatics tools in new ways. The future may not look at all like the past; we may now be able to make cheminformatics more accessible to the masses as it is essential to turn our accumulated data into something of real value that leads to biomedical advances. Our efforts in applying these various approaches to neglected diseases are just one example. That impact of cheminformatics in itself, is an accomplishment that is worthy of more support whether governmental or otherwise.
Acknowledgements
S.E. gratefully acknowledges colleagues at CDD, Dr. Joel S. Freundlich (Rutgers), Dr. Malabika Sarker (SRI) and Dr. Katalin Nadassy (Accelrys) for valuable discussions and assistance in developing some of the projects discussed. S.E. acknowledges that the Bayesian models were developed with support from Award Number R43 LM011152-01 “Biocomputation across distributed private datasets to enhance drug discovery” from the National Library of Medicine. TB Mobile and the associated datasets was made possible with funding from Award Number 2R42AI088893-02 “Identification of novel therapeutics for tuberculosis combining cheminformatics, diverse databases and logic based pathway analysis” from the National Institutes of Allergy and Infectious Diseases.
Footnotes
Competing Financial Interests
NL is an employee and SE is a consultant for CDD Inc. SE is on the advisory board for Assay Depot. AJW is an employee of the Royal Society of Chemistry. AMC is the founder of Molecular Materials Informatics and a consultant for CDD.
References
- 1.Villoutreix BO, Lagorce D, Labbe CM, Sperandio O, Miteva MA. One hundred thousand mouse clicks down the road: selected online resources supporting drug discovery collected over a decade. Drug Discov Today. 2013;18:1081–1089. doi: 10.1016/j.drudis.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Pence HE, Williams AJ. ChemSpider: An Online Chemical Information Resource. J Chem Educ. 2010;87:1123–1124. [Google Scholar]
- 3.Ekins S, Williams AJ. Precompetitive Preclinical ADME/Tox Data: Set It Free on The Web to Facilitate Computational Model Building to Assist Drug Development. Lab on a Chip. 2010;10:13–22. doi: 10.1039/b917760b. [DOI] [PubMed] [Google Scholar]
- 4.Ekins S, Andreyev S, Ryabov A, Kirillov E, Rakhmatulin EA, Bugrim A, Nikolskaya T. Computational prediction of human drug metabolism. Expert Opin Drug Metab Toxicol. 2005;1:303–324. doi: 10.1517/17425255.1.2.303. [DOI] [PubMed] [Google Scholar]
- 5.Ekins S, Andreyev S, Ryabov A, Kirillov E, Rakhmatulin EA, Sorokina S, Bugrim A, Nikolskaya T. A Combined Approach to Drug Metabolism and Toxicity Assessment. Drug Metab Dispos. 2006;34:495–503. doi: 10.1124/dmd.105.008458. [DOI] [PubMed] [Google Scholar]
- 6.Ekins S, Bugrim A, Brovold L, Kirillov E, Nikolsky Y, Rakhmatulin EA, Sorokina S, Ryabov A, Serebryiskaya T, Melnikov A, Metz J, Nikolskaya T. Algorithms for network analysis in systems-ADME/Tox using the MetaCore and MetaDrug platforms. Xenobiotica. 2006;36:877–901. doi: 10.1080/00498250600861660. [DOI] [PubMed] [Google Scholar]
- 7.Ekins S, Kirillov E, Rakhmatulin EA, Nikolskaya T. A Novel Method for Visualizing Nuclear Hormone Receptor Networks Relevant to Drug Metabolism. Drug Metab Dispos. 2005;33:474–481. doi: 10.1124/dmd.104.002717. [DOI] [PubMed] [Google Scholar]
- 8.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 9.Embrechts MJ, Ekins S. Classification of Metabolites with Kernel-Partial Least Squares (K-PLS). Drug Metab Dispos. 2007;35:325–327. doi: 10.1124/dmd.106.013185. [DOI] [PubMed] [Google Scholar]
- 10.Stranz DD, Miao S, Campbell S, Maydwell G, Ekins S. Combined computational metabolite prediction and automated structure-based analysis of mass spectrometric data. Toxicol Mech Methods. 2008;18:243–250. doi: 10.1080/15376510701857189. [DOI] [PubMed] [Google Scholar]
- 11.Hohman M, Gregory K, Chibale K, Smith PJ, Ekins S, Bunin B. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Disc Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Bost F, Jacobs RT, Kowalczyk P. Informatics for neglected diseases collaborations. Curr Opin Drug Discov Devel. 2010;13:286–296. [PubMed] [Google Scholar]
- 13.Bunin BA, Ekins S. Alternative business models for drug discovery. Drug Disc Today. 2011;16:643–645. doi: 10.1016/j.drudis.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Sarker M, Talcott C, Madrid P, Chopra S, Bunin BA, Lamichhane G, Freundlich JS, Ekins S. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium tuberculosis. Pharm Res. 2012;29:2115–2127. doi: 10.1007/s11095-012-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekins S, Hupcey MAZ, Williams AJ. Collaborative computational technologies for biomedical research. Wiley; Hoboken: NJ: 2011. [Google Scholar]
- 16.Ekins S, Hohman M, Bunin BA. In: Collaborative Computational Technologies for Biomedical Research. Ekins S, Hupcey MAZ, Williams AJ, editors. Wiley and Sons; Hoboken: 2011. [Google Scholar]
- 17.Burrill GS. 4th Annual CDD Community Meeting; San Francisco. 2010. [Google Scholar]
- 18.Todd MH. Open access and open source in chemistry. Chem Cent J. 2007;1:3. doi: 10.1186/1752-153X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardal C, Rottingen JA. Open source drug discovery in practice: a case study. PLoS Negl Trop Dis. 2012;6:e1827. doi: 10.1371/journal.pntd.0001827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anon European Lead Factory http://www.europeanleadfactory.eu/#.
- 21.Anon Elixir http://www.elixir-europe.org/
- 22.Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov Today. 2010;15:1052–1057. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams AJ, Ekins S. A quality alert and call for improved curation of public chemistry databases. Drug Disc Today. 2011;16:747–750. doi: 10.1016/j.drudis.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Williams AJ, Ekins S, Tkachenko V. Towards a Gold Standard: Regarding Quality in Public Domain Chemistry Databases and Approaches to Improving the Situation. Drug Disc Today. 2012;17:685–701. doi: 10.1016/j.drudis.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE, Sachs JD, Savioli L. Control of neglected tropical diseases. N Engl J Med. 2007;357:1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 26.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jimenez-Diaz MB, Martinez MS, Wilson EB, Tripathi AK, Gut J, Sharlow ER, Bathurst I, El Mazouni F, Fowble JW, Forquer I, McGinley PL, Castro S, Angulo-Barturen I, Ferrer S, Rosenthal PJ, Derisi JL, Sullivan DJ, Lazo JS, Roos DS, Riscoe MK, Phillips MA, Rathod PK, Van Voorhis WC, Avery VM, Guy RK. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro I, Sevcsik AM, Alves F, Diap G, Don R, Harhay MO, Chang S, Pecoul B. New, improved treatments for Chagas disease: from the R&D pipeline to the patients. PLoS Negl Trop Dis. 2009;3:e484. doi: 10.1371/journal.pntd.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bettiol E, Samanovic M, Murkin AS, Raper J, Buckner F, Rodriguez A. Identification of three classes of heteroaromatic compounds with activity against intracellular Trypanosoma cruzi by chemical library screening. PLoS Negl Trop Dis. 2009;3:e384. doi: 10.1371/journal.pntd.0000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponder EL, Freundlich JS, Sarker M, Ekins S. Computational Models for Neglected Diseases: Gaps and Opportunities. Pharm Res. 2014;31:271–277. doi: 10.1007/s11095-013-1170-9. [DOI] [PubMed] [Google Scholar]
- 30.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 31.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, Gonzalez R, Lozano S, Huss S, Santos-Villarejo A, Martin-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuinan-Blanco M, Lavandera JL, Perez-Herran E, Gamo-Benito FJ, Garcia-Bustos JF, Barros D, Castro JP, Cammack N. Fueling Open-Source Drug Discovery: 177 Small-Molecule Leads against Tuberculosis. ChemMedChem. 2013;8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamo F-J, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera J-L, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 33.Anon TB Alliance Preclinical Pipeline http://www.tballiance.org/downloads/Pipeline/TBA%20Pipeline%20Q1%202014%282%29%20%28DA%29.pdf.
- 34.Anon TB Alliance Clinical Pipeline http://www.tballiance.org/portfolio/
- 35.Ekins S, Pottorf R, Reynolds RC, Williams AJ, Clark AM, Freundlich JS. Looking Back To The Future: Predicting In vivo Efficacy of Small Molecules Versus Mycobacterium tuberculosis. J Chem Inf Model. 2014;54:1070–1082. doi: 10.1021/ci500077v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballell L, Field RA, Duncan K, Young RJ. New small-molecule synthetic antimycobacterials. Antimicrob Agents Chemother. 2005;49:2153–2163. doi: 10.1128/AAC.49.6.2153-2163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds RC, Ananthan S, Faaleolea E, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Sosa MI, Thammasuvimol E, White EL, Zhang W, Secrist JA., 3rd High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2011 doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddry JA, Ananthan S, Goldman RC, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Sosa MI, White EL, Zhang W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ananthan S, Faaleolea ER, Goldman RC, Hobrath JV, Kwong CD, Laughon BE, Maddry JA, Mehta A, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Shindo N, Showe DN, Sosa MI, Suling WJ, White EL. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekins S, Freundlich JS, Hobrath JV, White EL, Reynolds RC. Combining Computational Methods for Hit to Lead Optimization in Mycobacterium tuberculosis Drug Discovery. Pharm Res. 2014;31:414–435. doi: 10.1007/s11095-013-1172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds RC, Ananthan S, Faaleolea E, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Sosa MI, Thammasuvimol E, White EL, Zhang W, Secrist JA., 3rd High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash O, Ghosh I. Developing an antituberculosis compounds database and data mining in the search of a motif responsible for the activity of a diverse class of antituberculosis agents. J Chem Inf Model. 2006;46:17–23. doi: 10.1021/ci050115s. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Garcia A, Galvez J, de Julian-Ortiz JV, Garcia-Domenech R, Munoz C, Guna R, Borras R. Search of chemical scaffolds for novel antituberculosis agents. J Biomol Screen. 2005;10:206–214. doi: 10.1177/1087057104273486. [DOI] [PubMed] [Google Scholar]
- 44.Planche AS, Scotti MT, Lopez AG, de Paulo Emerenciano V, Perez EM, Uriarte E. Design of novel antituberculosis compounds using graph-theoretical and substructural approaches. Mol Divers. 2009;13:445–458. doi: 10.1007/s11030-009-9129-9. [DOI] [PubMed] [Google Scholar]
- 45.Sundaramurthi JC, Brindha S, Reddy TB, Hanna LE. Informatics resources for tuberculosis--towards drug discovery. Tuberculosis (Edinb) 2012;92:133–138. doi: 10.1016/j.tube.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Ekins S, Freundlich JS, Choi I, Sarker M, Talcott C. Computational Databases, Pathway and Cheminformatics Tools for Tuberculosis Drug Discovery. Trends in Microbiology. 2011;19:65–74. doi: 10.1016/j.tim.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ekins S, Freundlich JS. Computational models for tuberculosis drug discovery. Methods Mol Biol. 2013;993:245–262. doi: 10.1007/978-1-62703-342-8_16. [DOI] [PubMed] [Google Scholar]
- 48.Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 49.Ekins S, Freundlich JS. Validating new tuberculosis computational models with public whole cell screening aerobic activity datasets. Pharm Res. 2011;28:1859–1869. doi: 10.1007/s11095-011-0413-x. [DOI] [PubMed] [Google Scholar]
- 50.Ekins S, Kaneko T, Lipinksi CA, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Ernst S, Yang J, Goncharoff N, Hohman M, Bunin B. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis. Mol BioSyst. 2010;6:2316–2324. doi: 10.1039/c0mb00104j. [DOI] [PubMed] [Google Scholar]
- 51.Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PLOSONE. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ekins S, Reynolds R, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chem Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ekins S, Freundlich JS, Reynolds RC. Are bigger datasets better for machine learning? Fusing single-point and dual-event dose response data for Mycobacterium tuberculosis. 2014 doi: 10.1021/ci500264r. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson JW, Sarantakis D, Terpinski J, Kumar TR, Tsai HC, Kuo M, Ager AL, Jacobs WR, Jr., Schiehser GA, Ekins S, Sacchettini JC, Jacobus DP, Fidock DA, Freundlich JS. Novel diaryl ureas with efficacy in a mouse model of malaria. Bioorg Med Chem Lett. 2012;23:1022–1025. doi: 10.1016/j.bmcl.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvarez G, Martinez J, Aguirre-Lopez B, Cabrera N, Perez-Diaz L, Gomez-Puyou MT, Gomez-Puyou A, Perez-Montfort R, Garat B, Merlino A, Gonzalez M, Cerecetto H. New chemotypes as Trypanosoma cruzi triosephosphate isomerase inhibitors: a deeper insight into the mechanism of inhibition. J Enzyme Inhib Med Chem. 2012 doi: 10.3109/14756366.2013.765415. [DOI] [PubMed] [Google Scholar]
- 56.Pires DE, de Melo-Minardi RC, da Silveira CH, Campos FF, Meira W., Jr. aCSM: noise-free graph-based signatures to large-scale receptor-based ligand prediction. Bioinformatics. 2013;29:855–861. doi: 10.1093/bioinformatics/btt058. [DOI] [PubMed] [Google Scholar]
- 57.Gunatilleke SS, Calvet CM, Johnston JB, Chen CK, Erenburg G, Gut J, Engel JC, Ang KK, Mulvaney J, Chen S, Arkin MR, McKerrow JH, Podust LM. Diverse inhibitor chemotypes targeting Trypanosoma cruzi CYP51. PLoS Negl Trop Dis. 2012;6:e1736. doi: 10.1371/journal.pntd.0001736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schamberger J, Grimm M, Steinmeyer A, Hillisch A. Rendezvous in chemical space? Comparing the small molecule compound libraries of Bayer and Schering. Drug Discov Today. 2011;16:636–641. doi: 10.1016/j.drudis.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 59.Kogej T, Blomberg N, Greasley PJ, Mundt S, Vainio MJ, Schamberger J, Schmidt G, Huser J. Big pharma screening collections: more of the same or unique libraries? The AstraZeneca-Bayer Pharma AG case. Drug Discov Today. 2012 doi: 10.1016/j.drudis.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Tu M, Rai BK, Mathiowetz AM, Didiuk M, Pfefferkorn JA, Guzman-Perez A, Benbow J, Guimaraes CR, Mente S, Hayward MM, Liras S. Exploring aromatic chemical space with NEAT: novel and electronically equivalent aromatic template. J Chem Inf Model. 2012;52:1114–1123. doi: 10.1021/ci300031s. [DOI] [PubMed] [Google Scholar]
- 61.Posner BA, Xi H, Mills JE. Enhanced HTS hit selection via a local hit rate analysis. J Chem Inf Model. 2009;49:2202–2210. doi: 10.1021/ci900113d. [DOI] [PubMed] [Google Scholar]
- 62.Gunter B, Brideau C, Pikounis B, Liaw A. Statistical and graphical methods for quality control determination of high-throughput screening data. J Biomol Screen. 2003;8:624–633. doi: 10.1177/1087057103258284. [DOI] [PubMed] [Google Scholar]
- 63.Varin T, Gubler H, Parker CN, Zhang JH, Raman P, Ertl P, Schuffenhauer A. Compound set enrichment: a novel approach to analysis of primary HTS data. J Chem Inf Model. 2010;50:2067–2078. doi: 10.1021/ci100203e. [DOI] [PubMed] [Google Scholar]
- 64.Swamidass SJ, Calhoun BT, Bittker JA, Bodycombe NE, Clemons PA. Enhancing the rate of scaffold discovery with diversity-oriented prioritization. Bioinformatics. 2011;27:2271–2278. doi: 10.1093/bioinformatics/btr369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swamidass SJ, Calhoun BT, Bittker JA, Bodycombe NE, Clemons PA. Utility-aware screening with clique-oriented prioritization. J Chem Inf Model. 2011;52:29–37. doi: 10.1021/ci2003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swamidass SJ. Using economic optimization to design high-throughput screens. Future Med Chem. 2013;5:9–11. doi: 10.4155/fmc.12.186. [DOI] [PubMed] [Google Scholar]
- 67.Makarenkov V, Kevorkov D, Zentilli P, Gagarin A, Malo N, Nadon R. HTS-Corrector: software for the statistical analysis and correction of experimental high-throughput screening data. Bioinformatics. 2006;22:1408–1409. doi: 10.1093/bioinformatics/btl126. [DOI] [PubMed] [Google Scholar]
- 68.Makarenkov V, Zentilli P, Kevorkov D, Gagarin A, Malo N, Nadon R. An efficient method for the detection and elimination of systematic error in high-throughput screening. Bioinformatics. 2007;23:1648–1657. doi: 10.1093/bioinformatics/btm145. [DOI] [PubMed] [Google Scholar]
- 69.Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calhoun BT, Browning MR, Chen BR, Bittker JA, Swamidass SJ. Automatically detecting workflows in PubChem. J Biomol Screen. 2012;17:1071–1079. doi: 10.1177/1087057112449054. [DOI] [PubMed] [Google Scholar]
- 71.Browning MR, Calhoun BT, Swamidass SJ. Managing missing measurements in small-molecule screens. J Comput Aided Mol Des. 2013 doi: 10.1007/s10822-013-9642-x. [DOI] [PubMed] [Google Scholar]
- 72.Schuffenhauer A, Ertl P, Roggo S, Wetzel S, Koch MA, Waldmann H. The scaffold tree--visualization of the scaffold universe by hierarchical scaffold classification. J Chem Inf Model. 2007;47:47–58. doi: 10.1021/ci600338x. [DOI] [PubMed] [Google Scholar]
- 73.Dimova D, Wawer M, Wassermann AM, Bajorath J. Design of multitarget activity landscapes that capture hierarchical activity cliff distributions. J Chem Inf Model. 2011;51:258–266. doi: 10.1021/ci100477m. [DOI] [PubMed] [Google Scholar]
- 74.Wassermann AM, Bajorath J. Directed R-group combination graph: a methodology to uncover structure-activity relationship patterns in a series of analogues. J Med Chem. 2012;55:1215–1226. doi: 10.1021/jm201362h. [DOI] [PubMed] [Google Scholar]
- 75.Howells J, Gagliardi D, Malik K. Sourcing knowledge: R&D outsourcing in UK pharmaceuticals. Int J Tech Man. 2012;59:139–161. [Google Scholar]
- 76.Fox S, Farr-Jones S, Sopchak L, Boggs A, Nicely HW, Khoury R, Biros M. High-throughput screening: update on practices and success. J Biomol Screen. 2006;11:864–869. doi: 10.1177/1087057106292473. [DOI] [PubMed] [Google Scholar]
- 77.McGee Outsourcing and contract services. J Biomol Screen. 2012;17:1379–1381. doi: 10.1177/1087057113505963. [DOI] [PubMed] [Google Scholar]
- 78.Ekins S, Olechno J, Williams AJ. Dispensing processes impact apparent biological activity as determined by computational and statistical analyses. PLoS One. 2013;8:e62325. doi: 10.1371/journal.pone.0062325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bradley D. Share and share alike. Nat Rev Drug Discov. 2005;4:180. doi: 10.1038/nrd1683. [DOI] [PubMed] [Google Scholar]
- 80.Masek BB, Shen L, Smith KM, Pearlman RS. Sharing chemical information without sharing chemical structure. J Chem Inf Model. 2008;48:256–261. doi: 10.1021/ci600383v. [DOI] [PubMed] [Google Scholar]
- 81.Balaban A. Can topological indices transmit information on properties but not on structures? J Comp Aided Mol Des. 2005;19:651–660. doi: 10.1007/s10822-005-9010-6. [DOI] [PubMed] [Google Scholar]
- 82.Bologa C, Allu TK, Olah M, Kappler MA, Oprea TI. Descriptor collision and confusion: toward the design of descriptors to mask chemical structures. J Comput Aided Mol Des. 2005;19:625–635. doi: 10.1007/s10822-005-9020-4. [DOI] [PubMed] [Google Scholar]
- 83.Clement OO, Guner OF. Possibilities for transfer of relevant data without revealing structural information. J Comput Aided Mol Des. 2005;19:731–738. doi: 10.1007/s10822-005-9026-y. [DOI] [PubMed] [Google Scholar]
- 84.Filimonov D, Poroikov V. Why relevant chemical information cannot be exchanged without disclosing structures. J Comput Aided Mol Des. 2005;19:705–713. doi: 10.1007/s10822-005-9014-2. [DOI] [PubMed] [Google Scholar]
- 85.Kaiser D, Zdrazil B, Ecker GF. Similarity-based descriptors (SIBAR)--a tool for safe exchange of chemical information? J Comput Aided Mol Des. 2005;19:687–692. doi: 10.1007/s10822-005-9000-8. [DOI] [PubMed] [Google Scholar]
- 86.Trepalin S, Osadchiy N. The centroidal algorithm in molecular similarity and diversity calculations on confidential datasets. J Comput Aided Mol Des. 2005;19:715–729. doi: 10.1007/s10822-005-9023-1. [DOI] [PubMed] [Google Scholar]
- 87.Tetko IV, Abagyan R, Oprea TI. Surrogate data--a secure way to share corporate data. J Comput Aided Mol Des. 2005;19:749–764. doi: 10.1007/s10822-005-9013-3. [DOI] [PubMed] [Google Scholar]
- 88.Karr AF, Feng J, Lin X, Sanil AP, Young SS, Reiter JP. Secure analysis of distributed chemical databases without data integration. J Comput Aided Mol Des. 2005;19:739–747. doi: 10.1007/s10822-005-9011-5. [DOI] [PubMed] [Google Scholar]
- 89.Faulon JL, Brown WM, Martin S. Reverse engineering chemical structures from molecular descriptors: how many solutions? J Comput Aided Mol Des. 2005;19:637–650. doi: 10.1007/s10822-005-9007-1. [DOI] [PubMed] [Google Scholar]
- 90.Matlock M, Swamidass SJ. Sharing Chemical Relationships Does Not Reveal Structures. J Chem Inf Model. 2014;54:37–48. doi: 10.1021/ci400399a. [DOI] [PubMed] [Google Scholar]
- 91.Swamidass SJ, Matlock M, Rozenblit L. When should we share? Securely measuring he overlap between private datasets. 2013. Submitted. [DOI] [PMC free article] [PubMed]
- 92.Huang Y, Shen C, Evans D, Katz J, Shelat A. In: Information Systems Security. Jajodia S, Mazumdar C, editors. Springer Heidelberg; 2011. pp. 28–48. [Google Scholar]
- 93.Kuzu M, Kantarcioglu M, Durham EA, Toth C, Malin B. A practical approach to achieve private medical record linkage in light of public resources. J Am Med Inform Assoc. 2013;20:285–292. doi: 10.1136/amiajnl-2012-000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johnson SB, Whitney G, McAuliffe M, Wang H, McCreedy E, Rozenblit L, Evans CC. Using global unique identifiers to link autism collections. J Am Med Inform Assoc. 2010;17:689–695. doi: 10.1136/jamia.2009.002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Warner DJ, Griffen EJ, St-Gallay SA. WizePairZ: a novel algorithm to identify, encode, and exploit matched molecular pairs with unspecified cores in medicinal chemistry. J Chem Inf Model. 2010;50:1350–1357. doi: 10.1021/ci100084s. [DOI] [PubMed] [Google Scholar]
- 96.Anon Roche and AstraZeneca launch medicinal chemistry data-sharing consortium to further accelerate drug discovery. 2013 http://www.astrazeneca.com/Research/news/Article/260613-roche-and-astrazeneca-launch-medicinal-chemistry-datasha.
- 97.Zaretzki J, Matlock M, Swamidass SJ. XenoSite: accurately predicting CYP-mediated sites of metabolism with neural networks. J Chem Inf Model. 2013;53:3373–3383. doi: 10.1021/ci400518g. [DOI] [PubMed] [Google Scholar]
- 98.Ekins S. Progress in computational toxicology. J Pharmacol Toxicol Methods. 2014;69:115–140. doi: 10.1016/j.vascn.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 99.Aruoja V, Moosus M, Kahru A, Sihtmae M, Maran U. Measurement of baseline toxicity and QSAR analysis of 50 non-polar and 58 polar narcotic chemicals for the alga Pseudokirchneriella subcapitata. Chemosphere. 2014;96:23–32. doi: 10.1016/j.chemosphere.2013.06.088. [DOI] [PubMed] [Google Scholar]
- 100.Sushko I, Novotarskyi S, Korner R, Pandey AK, Rupp M, Teetz W, Brandmaier S, Abdelaziz A, Prokopenko VV, Tanchuk VY, Todeschini R, Varnek A, Marcou G, Ertl P, Potemkin V, Grishina M, Gasteiger J, Schwab C, Baskin II, Palyulin VA, Radchenko EV, Welsh WJ, Kholodovych V, Chekmarev D, Cherkasov A, Aires-de-Sousa J, Zhang QY, Bender A, Nigsch F, Patiny L, Williams A, Tkachenko V, Tetko IV. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J Comput Aided Mol Des. 2011;25:533–554. doi: 10.1007/s10822-011-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker T, Grulke CM, Pozefsky D, Tropsha A. Chembench: a cheminformatics workbench. Bioinformatics. 2010;26:3000–3001. doi: 10.1093/bioinformatics/btq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gupta RR, Gifford EM, Liston T, Waller CL, Bunin B, Ekins S. Using open source computational tools for predicting human metabolic stability and additional ADME/TOX properties. Drug Metab Dispos. 2010;38:2083–2090. doi: 10.1124/dmd.110.034918. [DOI] [PubMed] [Google Scholar]
- 103.Spjuth O, Willighagen EL, Guha R, Eklund M, Wikberg JE. Towards interoperable and reproducible QSAR analyses: Exchange of datasets. J Cheminform. 2010;2:5. doi: 10.1186/1758-2946-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams AJ, Ekins S, Spjuth O, Willighagen EL. Accessing, using, and creating chemical property databases for computational toxicology modeling. Methods Mol Biol. 2012;929:221–241. doi: 10.1007/978-1-62703-050-2_10. [DOI] [PubMed] [Google Scholar]
- 105.Ekins S, Gupta RR, Gifford E, Bunin BA, Waller CL. Chemical space: missing pieces in cheminformatics. Pharm Res. 2010;27:2035–2039. doi: 10.1007/s11095-010-0229-0. [DOI] [PubMed] [Google Scholar]
- 106.Guha R, Spjuth O, Willighagen EL. In: Collaborative computational technologies for biomedical research. Ekins S, Hupcey MAZ, Williams AJ, editors. Wiley and Sons; Hoboken: 2011. pp. 399–422. [Google Scholar]
- 107.Spjuth O, Carlsson L, Alvarsson J, Georgiev V, Willighagen E, Eklund M. Open source drug discovery with bioclipse. Curr Top Med Chem. 2012;12:1980–1986. doi: 10.2174/156802612804910287. [DOI] [PubMed] [Google Scholar]
- 108.Williams AJ, Harland L, Groth P, Pettifer S, Chichester C, Willighagen EL, Evelo CT, Blomberg N, Ecker G, Goble C, Mons B. Open PHACTS: Semantic interoperability for drug discovery. Drug Disc Today. 2012 doi: 10.1016/j.drudis.2012.05.016. In press. [DOI] [PubMed] [Google Scholar]
- 109.Ekins S, Bugrim A, Nikolsky Y, Nikolskaya T. In: Drug discovery handbook. Gad S, editor. Wiley; New York: 2005. pp. 123–183. [Google Scholar]
- 110.Anon ACS GCI Pharmaceutical Roundtable http://portal.acs.org/portal/acs/corg/content?_nfpb=true&_pageLabel=PP_TRANSITIONMAIN&node_id=1422&use_sec=false&sec_url_var=region1&__uuid=46aca9b6-a985-42cd-a534-7d6cabf892a7.
- 111.Ekins S, Clark AM, Williams AJ. Incorporating Green Chemistry Concepts into Mobile Chemistry Applications and Their Potential Uses. ACS Sustain Chem Eng. 2013;1:8–13. [Google Scholar]
- 112.Ekins S, Casey AC, Roberts D, Parish T, Bunin BA. Bayesian Models for Screening and TB Mobile for Target Inference with Mycobacterium tuberculosis. Tuberculosis (Edinb) 2013 doi: 10.1016/j.tube.2013.12.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ekins S, Clark AM, Sarker M. TB Mobile: A Mobile App for Anti-tuberculosis Molecules with Known Targets. J Cheminform. 2013;5:13. doi: 10.1186/1758-2946-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Clark AM, Sarker M, Ekins S. New target predictions and visualization tools incorporating open source molecular fingerprints for TB Mobile 2.0. submitted. 2014. [DOI] [PMC free article] [PubMed]
- 115.Ekins S, Clark AM, Williams AJ. Open Drug Discovery Teams: A Chemistry Mobile App for Collaboration. Mol Inform. 2012;31:585–597. doi: 10.1002/minf.201200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Clark AM, Ekins S, Williams AJ. Redefining cheminformatics with intuitive collaborative mobile apps. Molecular Informatics. 2012;31:569–584. doi: 10.1002/minf.201200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Clark AM, Williams AJ, Ekins S. Cheminformatics workflows using mobile apps. Chem-Bio Informatics J. 2013;13:1–18. [Google Scholar]
- 118.Ekins S, Waller CL, Bradley MP, Clark AM, Williams AJ. Four Disruptive Strategies for Removing Drug Discovery Bottlenecks. Drug Disc Today. 2013;18:265–271. doi: 10.1016/j.drudis.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 119.Anon Assay Depot https://www.assaydepot.com/
- 120.Anon Science Exchange https://www.scienceexchange.com/
- 121.Anon Euretos http://euretos.com/brain.
- 122.van Haagen HH, t Hoen PA, Mons B, Schultes EA. Generic information can retrieve known biological associations: implications for biomedical knowledge discovery. PLoS One. 2013;8:e78665. doi: 10.1371/journal.pone.0078665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ekins S, Polli JE, Swaan PW, Wright SH. Computational Modeling to Accelerate the Identification of Substrates and Inhibitors For Transporters That Affect Drug Disposition. Clin Pharmacol Ther. 2012;92:661–665. doi: 10.1038/clpt.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beaulieu CL, Ekins S, Samuels M, Boycott KM, MacKenzie A. Towards the development of a generalizable pre-clinical research pathway for orphan disease therapy. Orphanet J Rare Dis. 2012;7:39. doi: 10.1186/1750-1172-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wood J, Sames L, Moore A, Ekins S. Multifaceted roles of ultra-rare and rare disease patients/parents in drug discovery. Drug Discov Today. 2013;18:1043–1051. doi: 10.1016/j.drudis.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 126.Williams AJ, Wilbanks J, Ekins S. Why Open Drug Discovery Needs Four Simple Rules for Licensing Data and Models. PLoS Comput Biol. 2012;8:e1002706. doi: 10.1371/journal.pcbi.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Houlton S. Science at your fingerprints. Chemistry World 27th Feb. 2014 [Google Scholar]
- 128.Ekins S, Clark AM, Williams AJ. Open Drug Discovery Teams: A Chemistry Mobile App for Collaboration. Mol Inf. 2012;31:585–597. doi: 10.1002/minf.201200034. [DOI] [PMC free article] [PubMed] [Google Scholar]