Abstract
We investigated how the equine fetus prepares its pre-immune humoral repertoire for an imminent exposure to pathogens in the neonatal period, particularly how the primary hematopoietic organs are equipped to support B cell hematopoiesis and immunoglobulin (Ig) diversity. We demonstrated that the liver and the bone marrow at approximately 100 days of gestation (DG) are active sites of hematopoiesis based on the expression of signature mRNA (c-KIT, CD34, IL7R, CXCL12, IRF8, PU.1, PAX5, NOTCH1, GATA1, CEBPA) and protein markers (CD34, CD19, IgM, CD3, CD4, CD5, CD8, CD11b, CD172A) of hematopoietic development and leukocyte differentiation molecules, respectively. To verify Ig diversity achieved during the production of B cells, V(D)J segments were sequenced in primary lymphoid organs of the equine fetus and adult horse, revealing that similar heavy chain VDJ segments and CDR3 lengths were most frequently used independent of life stage. In contrast, different lambda light chain segments were predominant in equine fetal compared to adult stage and, surprisingly, the fetus had less restricted use of variable gene segments to construct the lambda chain. Fetal Igs also contained elements of sequence diversity, albeit to a smaller degree than that of the adult horse. Our data suggest that the B cells produced in the liver and bone marrow of the equine fetus generate a wide repertoire of pre-immune Igs for protection, and the more diverse use of different lambda variable gene segments in fetal life may provide the neonate an opportunity to respond to a wider range of antigens at birth.
Keywords: Equine, Hematopoiesis, B cell, Immunoglobulin, Diversity, Development
Introduction
Understanding the development of the immune system is critical for the development of successful vaccines against infectious agents that continue to cause significant disease in neonates and in the young. The purpose of our study was to learn how the liver and bone marrow of the equine fetus were equipped to support B cell hematopoiesis and immunoglobulin (Ig) diversity of the pre-immune repertoire. During fetal life, the liver is one primary hematopoietic organ and supports expansion of B cells until the bone marrow takes over this roll (Butler et al. 2011; Timens and Kamps 1997; Yokota et al. 2006). The horse is an ideal model to study the development of the humoral response during gestation, as the epitheliochorial placentation of the horse does not allow transfer of maternal immunoglobulins (Igs) to the fetus, eliminating this confounding element (Perryman et al. 1980).
B lymphopoiesis can be readily detected at the molecular level in the equine fetus around 90 to 120 days of gestation but perhaps even earlier in the yolk sac of the embryo (Tallmadge et al. 2009; Tallmadge et al. 2013; Tallmadge et al. 2014). Endogenous antibodies are first detected midway (around 180 days) through gestation and, when challenged in utero, the equine fetus generates an antigen-specific IgM and IgG antibody response by at least 200 days of gestation (DG) (Martin and Larson 1973; Morgan et al. 1975). Relatively little is known, however, about the generation of the immunoglobulin repertoire in the equine fetus, particularly with relevance to preparedness for fighting pathogens.
Critical to B lymphopoiesis is the generation of a functional Ig molecule, which requires somatic recombination of the V(D)J loci for both the heavy and light chain genes. The pre-immune Ig receptor repertoire develops in the absence of exogenous antigens in the primary lymphoid tissues, and diversity is generated primarily by combinatorial and junctional diversities. Combinatorial diversity is produced by combining different heavy chain and light chain gene segments. The number of gene segment used to construct Ig molecules varies by species: for example, the mouse has more than 90 functional IGHV segments while the chicken has only one (Das et al. 2008); the horse uses 14 IGHV, 40 IGHD, and 8 IGHJ functional segments to construct the heavy chain (Sun et al. 2010). Light chains are constructed using either the lambda or kappa loci. The horse has 27 IGLV, 7 IGLJ, and 7 IGLC potentially functional genes for the lambda light chain; and 19 IGKV, 4 IGKJ, and 1 IGKC for the kappa light chain (Sun et al. 2010). The Ig segments have been divided into subgroups, and each subgroup is composed of gene segments sharing >75% nucleotide identity (Sun et al. 2010). For the heavy chain, the 14 IGHV genes are grouped into 7 subgroups; the 40 IGHD genes into 28 subgroups; and the 8 IGHJ genes into 2 subgroups. The 27 IGLV genes were grouped into 11 subgroups. Recently, we proposed a change in nomenclature for the heavy chain Ig genes in accordance with the International ImMunoGeneTics information system based on these subgroups, and “VH5” was renamed “IGHV2S3”, indicating gene 3 of subgroup 2 (Tallmadge et al. 2013). Unlike the heavy chain nomenclature, the subgroup number is not followed by an “S” but a “-”, and the gene number based on chromosomal location from 3′ to 5′ in the locus (as annotated by Sun et al. 2010). Accordingly, “Vλ5” was renamed IGLV4-66, indicating subgroup 4 and the 66th IGLV gene from the 3′ end of the locus (Tallmadge et al. 2014). IGLJ genes names do not include a subgroup designation, and are named based on chromosomal location from 5′ to 3′ in the locus (Lefranc 2001).
The horse, sheep, cow, and chicken utilize the lambda light chain in more than 90% of circulating Ig molecules, in contrast to mice and rabbits with a predominance of the kappa chain, and humans and pigs with comparable use of lambda and kappa chains (Butler et al. 2006; Ford et al. 1994; Hood et al. 1967; Kelus and Weiss 1977). In some species, including humans (Zemlin et al. 2001), mice (Yancopoulos et al. 1984), pigs (Butler et al. 2000), cattle (Koti et al. 2010), sheep (Gontier et al. 2005), and zebrafish (Jiang et al. 2011), combinatorial diversity differs during phases of development, and certain Ig segments are preferentially utilized in fetal or adult life.
Junctional diversity is created by the deletion and addition of base pairs at the junctions of the Ig gene segments during recombination events. Addition of base pairs is created by the enzyme terminal deoxynucleotidyl transferase (TdT) that adds non-template nucleotides, or N-nucleotides, to the regions flanking Ig gene segments (Desiderio et al. 1984). Palindromic, or P-nucleotides, are template dependent dinucleotide additions palindromic to the flanking joining ends (Feeney 1990). Species with limited combinatorial diversity including swine (Butler et al. 2006; Butler et al. 2011), sheep (Reynaud et al. 1995), cattle (Kaushik et al. 2009; Koti et al. 2010; Saini and Kaushik 2002; Verma and Aitken 2012) depend more on junctional diversity and post-recombination processes such as somatic hypermutation. Chickens utilize somatic gene conversion (Reynaud et al. 1989), while rabbits use both somatic hypermutation and gene conversion to diversify their Ig repertoires (Becker and Knight 1990; Pinheiro et al. 2011).
The structure of the antibody molecule includes four framework sequences separated by three complimentary determining regions (CDRs). The CDRs are the sequences that come in contact with antigen, and are the most diverse regions of the Ig molecule. CDR3 contains the greatest degree of sequence variability and spans the junction of the IGHV, IGHD, and IGHJ gene segments in the heavy chain, and IGLV and IGLJ gene segments in the light chain. Diversity of the CDR3 region is influenced by both combinatorial and junctional diversity. CDR3 length differs in fetal compared to adult life in certain species including humans (Zemlin et al. 2001), mice (Bangs et al. 1991), and sheep (Gontier et al. 2005).
Our previous studies have described the generation of B cells and production of the Ig repertoire in the lymphoid tissues of the horse during phases of development (Tallmadge et al. 2009; Tallmadge et al. 2013; Tallmadge et al. 2014). We found that some aspects of Ig diversity were already established early in development while others appeared to be developmentally programed. For example, bias in IGHV segments present in the equine adult repertoire were also present in the fetal spleen (Tallmadge et al. 2013), while IGLV segment utilization was less restricted in the fetal compared to adult Ig repertoire (Tallmadge et al. 2014). However, it was unknown the degree of Ig diversity originated in the primary (liver and bone marrow) lymphoid tissues of the fetus, and further diversification in secondary lymphoid tissues (e.g. spleen). In this study, we tested how primary lymphoid tissues of the equine fetus were equipped to support B cell hematopoiesis and Ig diversity of the pre-immune repertoire during gestation. Our original hypothesis was that B cell hematopoiesis in the equine fetus occurs in the liver and bone marrow with limited Ig diversity. Herein we show, for the first time, active B cell hematopoiesis in the equine fetal liver and bone marrow around 100DG, with a surprising generation of a somewhat diverse pre-immune Ig repertoire that sets up humoral protection after birth.
Materials and methods
2.1 Equine tissue samples
These experiments were approved by the Cornell University Center for Animal Resources and Education and Institutional Animal Care and Use Committee for the use of vertebrates in research. Three mares (2 Thoroughbreds, 1 Warmblood) at the Cornell Equine Park were bred and abortions were chemically induced with prostaglandin injections (2.5mg intramuscularly every 12 hours until abortion, Lutalyse, Pfizer, New York, NY) at approximately 100DG (101-105DG) (Douglas and Ginther 1976). The fetal livers (n=3) were dissected out within an hour of abortion using sterile technique. Part of the tissues were snap frozen in liquid nitrogen for RNA isolation, or preserved in Tissue-Tek O.C.T. Compound (optimal tissue culture medium, SakurFineteck U.S.A., Inc., Torrance, CA) for immunohistochemistry; these tissues were stored at −80° until analysis. Single cell suspension was made from the remaining fetal liver by pressing the tissue through first a metal mesh (size 80 mesh, Sigma-Aldrich, St. Louis, MO) and then a nylon mesh (70μm, BD Falcon, Franklin Lakes, NJ). Mononuclear leukocytes were isolated by Ficoll gradient centrifugation (density 1.077, GE Healthcare, Piscataway, NJ) as previously described (Flaminio et al. 2000). Cells were immediately used for flow cytometric analysis or frozen in media containing dimethyl sulfoxide and stored in liquid nitrogen until additional flow cytometric analysis (CD5, CD34) or RNA isolation. Upon thawing for flow cytometric analysis, the cells were again subjected to Ficoll gradient centrifugation to enrich for viable cells. Fetal bone marrow was harvested from the same fetuses and snap frozen in liquid nitrogen for RNA isolation, and stored at −80°C until use. The adult horse bone marrow samples were collected immediately post-mortem from three healthy research adult horses (1 Thoroughbred, 2 breed unknown) belonging to another investigation at Cornell University College of Veterinary Medicine. The tissue was harvested within 1 hour of euthanasia, snap frozen in liquid nitrogen for RNA isolation, and stored at −80°C until use.
2.2 RT-PCR to detect expression of genes associated with hematopoiesis
Snap frozen equine fetal whole liver, fetal whole bone marrow, adult whole bone marrow tissue, and frozen fetal liver isolated leukocytes were homogenized with the QIAshredder columns (Qiagen, Valencia, CA). Total RNA was isolated with the RNeasy® Kit (Qiagen) and genomic DNA was degraded with the RNase-Free DNase Set® (Qiagen) or DNAse I (Life Technologies, Grand Island, NY) following the manufacturer’s instructions. cDNA was made from 1μg RNA with the RevertAID™ first strand cDNA synthesis kit using Oligo(dT)18 (Thermo Fisher Scientific Inc., Waltham, MA) according to manufacturer’s instructions. PCR reactions were performed by adding 50ng cDNA template to 0.2mM dNTP, 0.6μM each forward and reverse primers (Supplemental Table 1), and 1.25u DreamTaq™ DNA polymerase (Thermo Fisher Scientific Inc.) in 1× DreamTaq™ buffer. Thermal cycling conditions were as follows: initial denaturation 95°C × 1min, [denaturation 95°C × 30s, annealing 58°C × 30s, extension 72°C × 1min] repeated 40 times, and final extension 72°C × 5min. Amplicons were visualized using electrophoresis on a 1% agarose gel stained with GelGreen nucleic acid stain (Phenix Research Products, Candler, NC) and the Gel Doc™ EZ Imager (Bio-Rad Laboratories Inc., Hercules, CA).
A panel of 12 genes known to participate in hematopoiesis were tested, including transcription factors essential for commitment and differentiation of the hematopoietic lineages: (1) GATA1 for erythropoiesis (Tsiftsoglou et al. 2009); (2) CEBPA for myelopoiesis (Koschmieder et al. 2009); (3) PAX5 for B cell lymphopoiesis (Cobaleda et al. 2007; Pridans et al. 2008); (4) IFR8 and (5) PU.1 for both myelopoiesis and B lymphopoiesis, with PU.1 having additional functions in erythropoiesis (Kastner and Chan 2008; Wang and Morse 2009); (6) NOTCH1 receptor essential for T cell commitment (Rothenberg 2011); HSC markers (7) CD34, a sialomucin important in HSC migration and homing (Nielsen and McNagny 2009), and (8) KIT, a transmembrane receptor essential for HSC signaling (Kent et al. 2008), also expressed during B cell ontogeny from the lymphoid-primed multipotent progenitor through common lymphoid progenitor (CLP) and pro-B cell stages, and acts synergistically with IL-7 to promote proliferation and differentiation of pro-B cells (McNiece et al. 1991); (9) IL7R expressed by the CLP through the early pre-B cell stage and signaling promotes survival, proliferation, and differentiation (Fry and Mackall 2002); (10) IgM expressed only by immature and mature B cells; the chemokine receptor (11) CXCL12 and (12) IL-7 expressed by microenvironment stromal cells to support hematopoiesis (Nagasawa 2006), for the homing of HSCs to the fetal liver and bone marrow, and the generation of pre-pro and pro-B cells (Nagasawa et al. 1996). The house-keeping gene β-ACTIN was used as a positive control. Primers (Supplemental Table 1) were designed using the Primer3 program accessible at http://frodo.wi.mit.edu/ (Rozen and Skaletsky 2000). To prevent amplification of genomic DNA, the primers spanned introns when possible. Destruction of genomic DNA from RNA samples was confirmed by performing the cDNA synthesis reaction without the addition of the RevertAID™ Reverse Transcriptase (Thermo Fisher Scientific Inc.) followed by the PCR reaction with the β-ACTIN primers to show no genomic DNA product was amplified. The amplicons were confirmed by sequencing.
2.3 Immunohistochemistry of leukocytes in the equine fetal liver
Whole fetal liver harvested from two different fetuses and frozen in Tissue-Tek O.C.T. were cut into 7μm thick sections on a microtome and fixed with acetone for 10 minutes. The sections were stained as previously reported (Tallmadge et al. 2009) using the following primary antibodies (Kydd et al. 1994; Lunn et al. 1998; Parrish et al. 1982): CD2 (clone HB88A, VMRD, Pullman, WA), CD3 (F6G.3(G12), J. Stott, University of California, Davis, CA), CD4 (HB61A, VMRD), CD5 (HT23A, VMRD), CD8 (HT14A, VMRD), CD19-like (cz2.1, D.F. Antczak, Cornell University, Ithaca, NY), IgM (CM7, AbDSerotec, Raleigh, NC), CD172A (IGHD59B, VMRD), major histocompatibility complex (MHC) class I (cz3, D.F. Antczak), MHC class II (cz11, D.F. Antczak), anti-canine parvovirus (negative control, CPV12, C. Parrish, Ithaca, NY). Images of the slides were taken with an Olympus BX-50 microscope, visualized with Metamorph software (Molecular Devices, Sunnyvale, California), and representative images were selected from both fetuses.
2.4 Flow cytometric immunophenotyping of the equine fetal liver isolated leukocytes
One million cells per sample were blocked with 10% normal goat serum and washed with PBS, then labeled with the same antibodies as above for the immunohistochemistry including CD2, CD3, CD4, CD5, CD8, CD19-like, IgM, CD172A, MHC class I, MHC class II, in addition to CD11b-FITC (M1/70.15.11.5, Miltenyi Biotec, Auburn, CA) and CD34 (1H6, R&D Systems, Minneapolis, MN). Cells stained with unconjugated primary antibodies were stained with goat anti-mouse IgG(H+L)-FITC (Jackson ImmunoResearch Laboratories, West Grove, PA). Finally the cells were fixed with 2% paraformaldehyde in phosphate buffered solution, and fluorescence was measured with the BD FACScalibur flow cytometer using an argon laser (Bio-Rad Laboratories Inc.). The negative control were cells stained only with the goat anti-mouse IgG(H+L)-FITC secondary antibody. One hundred thousand ungated events were collected in all but one case (CD5 staining of fetal liver #1, 68,000 events counted).
2.5 Cytology of the equine fetal liver isolated leukocytes
Cytospins of approximately 5×104 isolated cells per slide were made using centrifugation at 500×g for 3 minutes, and stained with Wright’s stain in an automated stainer (Modified Wright’s stain, Hema-tek 1000, Siemens Healthcare Diagnostics Inc., Tarrytown, NJ) at the Clinical Pathology Laboratory, Cornell University College of Veterinary Medicine. A 200 cell differential count was performed by a single blinded observer (TS).
2.6 Immunoglobulin heavy and light chain V(D)J sequencing
RNA was isolated from whole tissues and genomic DNA destroyed as described above. 5′-rapid amplification of cDNA ends (RACE) library was constructed with the SMARTer™ RACE cDNA Amplification Kit (Clontech, Mountain View, CA) following manufacturer’s instructions. The 5′-RACE PCR was performed with 5′ RACE primers for the heavy chain VDJ sequence that spans the last 24 nucleotides conserved among IGHJ segments, and 5′ RACE primers lambda light chain sequence that spans a conserved sequence in the lambda constant segments, as previously described (Tallmadge et al. 2013; Tallmadge et al. 2014). The lambda chain was chosen as the horse uses primarily this light chain in circulating antibodies (Ford et al. 1994). The PCR products were purified, ligated into the pJET1.2 vector (CloneJET™ PCR Cloning Kit, Thermo Fisher Scientific Inc.), transformed into either JM107 (TransformAID™ Bacterial Transformation Kit, Thermo Fisher Scientific Inc.) or NEB-5 alpha (New England BioLabs, Ipswich, MA) competent E.coli, expanded, and sequenced at the Cornell University Institute of Biotechnology (Ithaca, NY), as previously described (Tallmadge et al. 2013; Tallmadge et al. 2014). Heavy and lambda light chain Ig sequences were obtained from fetal liver, fetal bone marrow, and adult bone marrow (n = 3). To recover a minimum of 30 unique and productive Ig sequences per tissue (10 per individual), 31 fetal liver, 49 fetal bone marrow, 32 adult horse bone marrow heavy chain clones and 33 fetal liver, 49 fetal bone marrow, and 33 adult horse bone marrow lambda light chain clones were sequenced. Minimal lambda light chain sequence diversity was obtained from donor fetal liver #2, despite sequencing 11 clones, resulting in only 2 unique clones from this donor.
2.7 Immunoglobulin heavy and lambda light chain sequence analysis
Ig sequences determined in this study are available through GenBank with accession numbers KF748612 - KF748792. Ig sequences were analyzed and nucleotide identity plots generated using Geneious Pro R6-1 (Drummond et al. 2011), (Biomatters Ltd, Auckland, New Zealand). Ig gene segments were identified by comparing the cloned sequences against the EquCab2.0 equine reference genome annotated by Sun et al. (2010) using the NCBI Equus caballus BLAST tools as previously described (Tallmadge et al. 2013; Tallmadge et al. 2014). All BLAST hits were evaluated for identity, alignment length, and orientation. With one exception (IGVDJ66), all annotated IGHD segments were at least 7bp long and shared greater than 65% nucleotide identity with the genomic sequence. In total, IGHD segments could be annotated in 86% of sequences. For the remainder of sequences with IGHD segments of insufficient length or nucleotide identity, IGHD segments were designated as “not determined”.
Ig gene sequence identities between expressed sequences and the genome reference sequences were calculated with the Geneious Pro R6-1 software. The length of the heavy chain (CDR3H) and lambda light chain (CDR3L) were determined as previously described (Ford et al. 1994; Sun et al. 2010). Variability plots were made as described by Wu and Kabat (1970) with the variability index calculated as the number of different amino acids at a given position divided by the frequency of the most common amino acid at that position.
2.8 Statistical analysis
The Shapiro-Wilk normality test performed with Graphpad Prism version 6.0c (GraphPad Software, San Diego, California) revealed that most of the data was not normally distributed, and the appropriate non-parametric test was performed. Pairwise nucleotide identity, nucleotide identity to genome, number of N-nucleotide additions, nucleotide deletions at segment junctions, IGHD segment length, and CDR3 lengths were evaluated with the Kruskal-Wallis Rank Sum test for three way comparisons between fetal liver, fetal bone marrow, and adult horse bone marrow, and the Wilcoxon-Mann-Whitney Rank Sum test for two-way comparisons between the different life stages or tissue with KaleidaGraph (Synergy Software, Reading, PA). IGHV, IGHD, and IGHJ segment usage was assessed by Chi2 analysis (Graphpad Prism version 6.0c). The Chi2 test was not valid for pairwise comparisons between tissues for IGLV segment use due to the use of many different gene segments resulting in a small frequency for any individual gene; therefore, the Fisher exact test was performed using Graphpad Prism for IGL segment usage. All data was treated as unpaired. A p-value ≤ 0.05 was considered significant.
Results
3.1 Molecular evidence of hematopoiesis
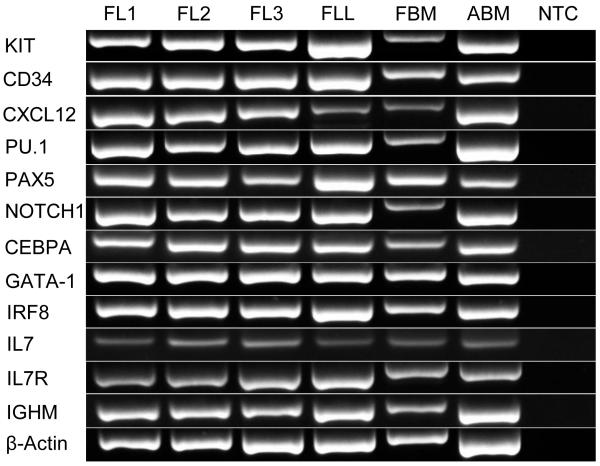
In order to characterize the fetal liver and bone marrow as active hematopoietic sites early in gestation, we confirmed the expression of relevant hematopoietic developmental genes. Twelve selected genes important in hematopoiesis KIT, CD34, IL7R, IGHM, CXCL12, IL7, PU.1, IRF8, PAX5, NOTCH1, CEPBA, and GATA1 were detected at the mRNA level in the adult horse bone marrow, and 100-day equine fetal liver and bone marrow whole tissues (Fig. 1). The same hematopoiesis-related genes were expressed in the isolated mononuclear cells from these tissues. The fetal bone marrow PCR products consistently ran slower than those from the other tissues, and direct sequencing of a subset of PCR reactions confirmed the bands were the amplicons of interest.
Fig. 1. Expression of hematopoietic genes in the equine fetal liver.
RNA was isolated from equine fetal liver and bone marrow collected at 100 days of gestation, and from adult horse bone marrow for expression analysis of the early genes in hematopoiesis using RT-PCR. FL1 = fetal liver 1, FL2 = fetal liver 2, FL3 = fetal liver 3, FLL = fetal liver leukocytes, FBM = fetal bone marrow, ABM = adult horse bone marrow, NTC = no template control.
3.2. Distribution of leukocytes in the fetal liver
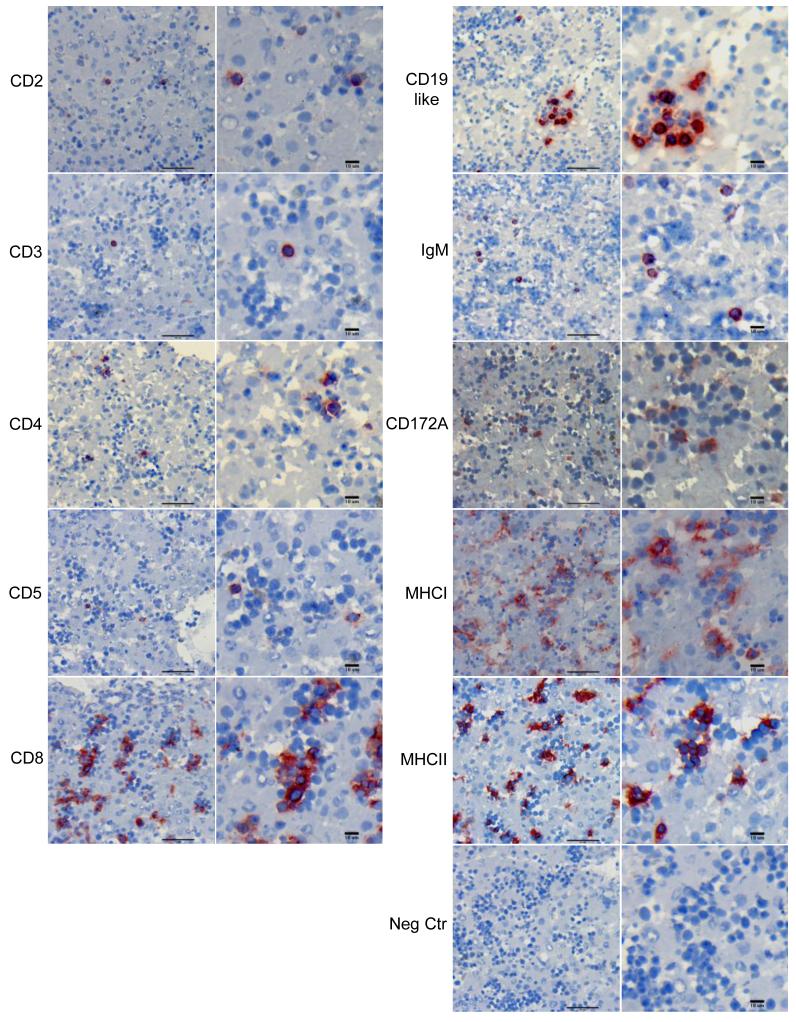
We next investigated how leukocytes were organized in the fetal liver, and in what proportion they were present in order to understand the relative extent of B cell hematopoiesis. Immunohistochemistry staining was used for qualitative analysis and showed the presence of leukocytes in the equine fetal liver at 100DG (Fig. 2). T cell (CD3+, CD4+, CD8+) and B cell (CD19+ or IgM+) markers stained cells with large nuclear to cytoplasmic ratios, consistent with the morphology of lymphocytes. These cells were randomly distributed throughout the tissue sections as isolated cells or in small clusters. CD2 and CD5, which are expressed by subsets of both T and B cell lymphocytes, stained cells in a similar distribution. The T cell marker CD8 stained a greater number of cells compared to the other T cell markers, and positive cells resembled lymphocytes and cells characteristic of the myeloid lineage, with small nuclear to cytoplasmic ratios. The CD172A marker, expressed by monocytes, macrophages and neutrophils, stained large and small cells with small nuclear to cytoplasmic ratios, and were distributed in small clusters. Cells staining positive for MHC class I and MHC class II were randomly distributed throughout the tissue. MHC class I positive cells were most often found in large groups (>10 cells), while MHC class II positive cells were identified generally in small groups or isolated. Similar to CD8, MHC class I and MHC class II positive cells resembled both the lymphocyte and myeloid lineages.
Fig. 2. Leukocyte distribution in the equine fetal liver.
Whole fresh-frozen equine fetal liver was stained with monoclonal antibodies against equine leukocyte markers, and representative images were selected from fetuses #2 and #3. The figures on the right of each panel show positive cells at higher magnification of the corresponding image on the left. Scale bars measure 50μm on the left column, and 10μm on the right column for each panel.
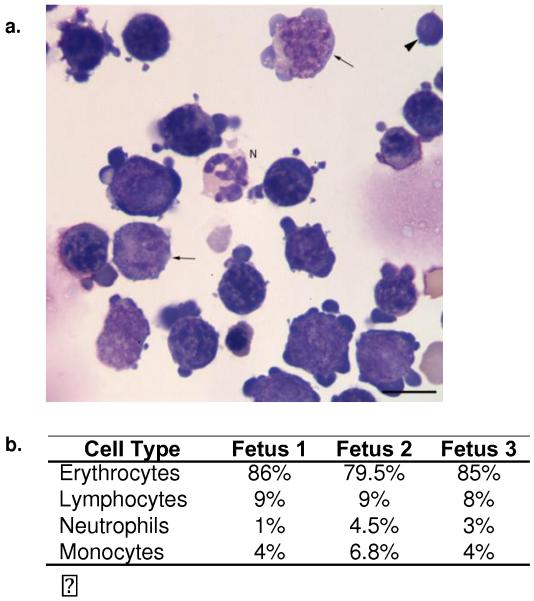
Isolated leukocytes from fetal liver were quantitated with flow cytometric analysis (Table 1). Approximately 2% of the total mononuclear cells isolated from the fetal liver expressed the HSC marker CD34, between 0.4-4.1% T or B cell markers, and 11-15% expressed the CD172A myeloid marker. The technique applied for leukocyte isolation (Ficoll gradient centrifugation) favors mononuclear cell enrichment, but not purification from neutrophils. In contrast, only 3% of cells stained positive with CD11b, which is expressed in monocytes, neutrophils, and B1 cells (Montecino-Rodriguez and Dorshkind 2006). Consistent with the immunohistochemistry staining, 24-30% cells expressed MHC class I, and 13-24% cells expressed MHC class II, the latter was similar to the percentage of B cells and monocytes together. Since only 12-19% of the cells isolated from the fetal liver could be accounted for as lymphoid or myeloid using the antibodies available for the horse, which do not include an erythrocyte lineage marker, we also classified the cells based on morphology using cytospin preparations. The differential cell count (Fig. 3) showed that the majority of cells (80-86%) were of the erythroid lineage, followed by cells with morphologic features of small lymphocytes (8-9%), monocytes (4-7%), and neutrophils (1-5%). The predominance of erythroid cells is consistent with the fetal liver’s function at this phase of life.
Table 1.
Surface molecule expression of equine fetal liver isolated leukocytes
| Percent Positive Cells Measured with Flow Cytometry | ||||
|---|---|---|---|---|
| Markers | Fetus 1 | Fetus 2 | Fetus 3 | Cells that express marker |
| CD2 | 1.9 | 2.1 | 2.0 | T cells, subset of B cells |
| CD3 | 0.8 | 0.9 | 1.3 | T cells |
| CD4 | 1.9 | 1.4 | 2.7 | T cells, subset of macrophages or dendritic cells |
| CD5 | 3.5 | 1.5 | 2.4 | T cells, subset of B cells |
| CD8 | 4.1 | 1.0 | 3.0 | T cells, subset of macrophages or dendritic cells |
| CD19-like | 1.0 | 0.4 | 1.6 | B cells |
| IgM | 0.9 | 0.5 | 1.4 | B cells |
| CD11b | 2.6 | NM | 3.2 | monocytes, macrophages, neutrophils, subset of B cells |
| CD172A | 15.5 | 11.8 | 14.4 | macrophages, neutrophils |
| CD34 | 2.3 | NM | 1.4 | hematopoietic stem cells |
| MHCI | 29.8 | 24.5 | 24.7 | mature nucleated cells |
| MHCII | 20.8 | 13.7 | 23.8 | monocytes, macrophages, dendritic cells, B cells |
‘NM’ indicates that the surface expression of the marker was not measured
Fig. 3. Leukocyte differential cell count in the equine fetal liver.
(a) Cytospin slides of isolated equine fetal liver leukocytes were made and stained with Wright’s stain. A sample image of fetus #3 is shown. Most of the nucleated cells were erythroid progenitors, with fewer differentiating monocytes (arrows), neutrophils (N) and lymphocytes (arrowhead). The scale bar measures 10μm. (b) Differential cell counts from each fetus sample show that the majority of cells were erythrocytes, followed by lymphocytes, monocytes, and neutrophils.
3.3 Sequence diversity in the pre-immune immunoglobulin repertoire
Previous studies showed that a small degree of sequence diversity was present in Ig derived from equine fetal spleen (Tallmadge et al. 2013; Tallmadge et al. 2014). In this part of our experiments, we asked if sequence diversity originated already in the primary lymphoid tissues of the fetus, and how that compared to the sequence diversity observed in the primary lymphoid tissue of the adult horse. Heavy and lambda light chain sequence variation was determined for equine fetal liver, fetal bone marrow, and adult horse bone marrow. The median percent nucleotide identity of heavy chain VDJ segments was compared between all tissues. Significant differences (p-values < 0.0001) were found between the fetal and adult horse tissues (Supplemental Fig. 1). In contrast to the heavy chain, the lambda light chain VJ segments had similar pairwise nucleotide identities between fetal and adult horse tissues (p-values > 0.16) (Supplemental Fig. 1).
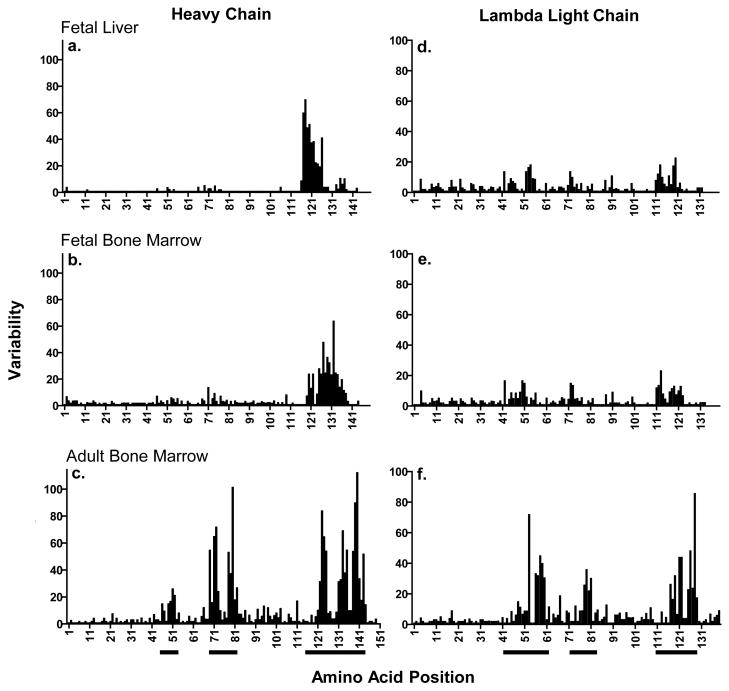
In the fetal tissues, these nucleotide differences were predominately in the CDR3H region (Supplemental Fig. 2), largely reflecting the use of different IGHD segments resulting in the greatest variability of amino acid identity in the positions spanning the CDR3H (Fig. 4). In the adult horse, the mutations and amino acid variability were concentrated in the CDR3H but also CDR2H, and to a lesser degree, CDR1H region (Supplemental Fig. 2 and Fig. 4). The amino acid differences in the lambda fetal sequences were more evenly distributed throughout the variable region with a smaller degree of clustering around the CDRs (Fig. 4 and Supplemental Fig. 3) than the fetal heavy chain sequences. In the adult horse bone marrow, however, the highest degree of amino acid variability was concentrated in the CDRs (Fig. 4).
Fig. 4. Variability in immunoglobulin amino acid sequence in equine fetus and adult horse primary lymphoid tissues.
Heavy and light chain Ig variable region nucleotide sequences, and their respective translated amino acid sequences were obtained from equine fetal (liver and bone marrow) and adult horse (bone marrow) whole tissues. Amino acid variability at each residue of the Ig variable region was calculated as describe by Wu and Kabat (1970) for (a) fetal liver heavy chain, (b) fetal bone marrow heavy chain, (c) adult horse bone marrow heavy chain, (d) fetal liver lambda light chain, (e) fetal bone marrow lambda light chain, and (f) adult horse bone marrow lambda light chain. The three CDR regions are underlined bellow the X-axis.
Fetal heavy chain sequences also had a higher median percentage (> 98%) of identical nucleotides with the reference genome sequence than sequences from horse bone marrow (89.4% for IGHV, 88.7% for IGHD, and 89.8% for IGHJ), and these germline identity values were significantly different (p-values < 0.03); germline identity values did not differ (p-values > 0.4) between fetal liver and fetal bone marrow (Supplemental Table 2). Similar to the heavy chain, lambda chain segments sequenced from equine fetal tissues (>97.3%) had higher median nucleotide identity to the reference genome sequences than the adult horse (88.1 % for IGLV, 91.7% for IGLJ), and these medians were significantly different (p-values <0.0001); germline identity values did not differ (p-values >0.4) between fetal liver and fetal bone marrow (Supplemental Table 3). Consistent with somatic hypermutation, adult horse sequences had higher mutation rates at adenosine nucleotides, and greater percentage of transition mutations for IGHV2S3 (35% in fetal verses 47% in adult horse), and IGLV8-128, IGLV8-24, IGLV8-12 (40% in fetal verses 45% in adult horse) (Di Noia and Neuberger 2007; Tonegawa 1983).
3.4 Combinatorial diversity in the pre-immune repertoire
The next question related to the extent of combinatorial diversity generated in the primary lymphoid tissues, and how it compared to secondary lymphoid tissues of the equine fetus or the adult. The Ig repertoire of the equine fetal liver and bone marrow showed similarities to that of the adult horse bone marrow, particularly in regards to combinatorial diversity of the heavy chain. IGHV2S2, IGHV2S3, and IGHV2S4 were observed in all tissues tested, all of which belong to IGHV Subgroup 2 as indicated by “2S” in the segment names, meaning that these IGHV germline genes share at least 75% identity at the nucleotide level to each other (Supplemental Fig. 4a; Sun et al. 2010; Tallmadge et al. 2013). Additionally IGHV segments from subgroups 1 and 4 were identified in the fetal bone marrow. IGHJ1S2, IGHJ1S3, and IGHJ1S5 were observed at all life stages with IGHJ1S4, IGHJ1S6, and IGHJ1S7 found in the fetal and adult horse bone marrow (Supplemental Fig. 4b). Twenty-three of the forty IGHD genes were annotated including 10 different IGHD segments in the fetal liver, 12 in the fetal bone marrow, and 15 in the adult horse bone marrow (Supplemental Fig. 4c). IGHD17S1 and IGHD17S2 share identical coding sequences and could not be distinguished from each other. There was one potential IGHD-IGHD rearrangement observed in IGVDJ75 in which there was a second IGHD segment that aligned between the first IGHD and IGHJ with junctional diversity on both the 5′ and 3′ junctions. However, only short 8-9 nucleotide sequences of both IGHD segments could be aligned and, therefore, the second shorter IGHD segment could not be confidently annotated. IGHV2S3, IGHJ1S5, IGHD18S1 were the most frequently utilized gene segments in fetal and adult horse tissues as previously reported (Sun et al. 2010; Tallmadge et al. 2013). When the composition of the most commonly used VDJ segments were compared, there was no statistically significant difference (p-value > 0.1) in usage of IGHV and IGHD segments between fetal liver and fetal bone marrow or between fetal and adult horse sequences; the difference in IGHJ utilization was significant only between fetal bone marrow and adult horse bone marrow but not between fetal bone marrow and fetal liver or any other comparison between fetal and adult horse sequences, making the biologic importance of this finding likely unimportant.
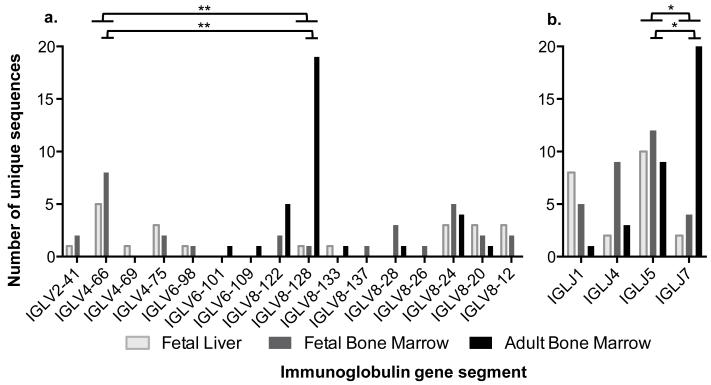
Sixteen different IGLV segments were observed (Fig. 5a) belonging to 4 different subgroups (Supplemental Table 3). Segments belonging to subgroups IGLV2 and IGLV4 were only isolated from fetal tissues, while those in IGLV6 and IGLV8 were found in all tissues. Subgroup IGLV2 consists of only one gene segment (IGLV2-41), while subgroups IGLV4, IGLV6, and IGLV8 have multiple gene segments (Sun et al. 2010). The four IGLJ gene segments identified were observed in all tissues tested (Fig. 5b). Fetus #2 had limited diversity with only two unique Ig lambda sequences collected, despite sequencing 11 clones. To make sure that the 5′ RACE library for this tissue was not biased, a second library was constructed, the lambda 5′RACE PCR repeated, and the resulting amplicons directly sequenced. The sequencing trace obtained was consistent with the clones already in hand so additional sequences were not pursued. IGLV4-66 and IGLJ5 were the most common segments used in fetal tissues, while IGLV8-128 and IGLJ7 were more frequently found in the adult horse bone marrow; this bias in IGLV and IGLJ segment use between fetal and adult horse sequences was statistically significant (p-values ≤ 0.0001 IGLV and ≤ 0.01 IGLJ, respectively) (Fig. 5).
Fig. 5. Immunoglobulin lambda light chain segment usage in equine fetal and adult horse primary lymphoid tissues.
(a) IGLV and (b) IGLJ gene segments were studied in the fetal liver (light gray), fetal bone marrow (dark gray), and adult horse bone marrow (black). The quantity of unique sequences utilizing each gene segment is shown. Significant differences (p-values ≤ 0.0001**) between the two most common IGLV segments (IGLV4-66, IGLV8-128) were observed with the Fisher’s Exact Test between fetal liver and adult horse bone marrow, fetal bone marrow and adult horse bone marrow, and all fetal and adult horse bone marrow; fetal liver and fetal bone marrow were not significantly different (p-value = 1.0). Significant differences (p-values ≤ 0.01*) were also observed for the two most common IGLJ (IGLJ5, IGLJ7) segments between fetal liver and adult horse bone marrow, fetal bone marrow and adult horse bone marrow, and all fetal and adult horse bone marrow; fetal liver and fetal bone marrow gene segment usages were not significantly different (p-value = 0.7).
A range of 14-28 unique heavy and lambda light chain Ig gene combinations were observed in fetal and adult horse tissues (Supplemental Fig. 5). The most frequently used heavy chain and light chain segments were not necessarily the most frequent combinations; for example, the most frequently used IGH segments were IGHV2S3, IGHJ1S5, IGHD18S1 but the most frequent combination in the fetal bone marrow was IGHV2S3-IGHD22S1-IGHJ1S5 (Supplemental Table 2). Similarly the most frequently observed IGL segments were IGLV4-66 and IGLJ5 in the fetus while the most frequent combination was IGLV4-66 -IGLJ1 in the fetal liver (Supplemental Table 3). Altogether, these data indicate that there is bias in variable region segment use in the lambda light chain but not the heavy chain for both life stages as previously reported in secondary lymphoid tissue (Tallmadge et al 2013; Tallmadge et al. 2014), and suggests that the heavy and lambda light chain rearrangements are regulated differently in primary fetal lymphoid tissue.
3.5 Junctional diversity in the pre-immune immunoglobulin repertoire
The 100DG equine fetus also had evidence of junctional diversity in the Ig heavy chain sequences, created by the deletion and addition of nucleotides at the V(D)J gene segment junctions. Non-template (N) nucleotides were present in fetal heavy chain sequences with medians of 7 (liver, range 0-35) and 5 (bone marrow, range 0-17) nucleotides at the IGHV-IGHD junction, and 0 nucleotides (liver range 0-9, bone marrow range 0-14) at the IGHD-IGHJ junction, similarly to secondary lymphoid tissue at this stage of development (Tallmadge et al. 2013). A significantly (p-values < 0.03) greater number of N-nucleotides were observed in adult horse heavy chain sequences with a median of 13 nucleotides (range 0-33) at the IGHV-IGHD, and 6 (range 0-50) at the IGHD-IGHJ junction when compared to both fetal liver and fetal bone marrow. There was no significantly statistical difference (p-values >0.4) in the number of N-nucleotides in the fetal liver compared to fetal bone marrow at any junction. A significantly (p-value <0.0001) greater number of N-nucleotides were also observed at the lambda chain IGLV-IGLJ junction in the adult horse bone marrow with a median of 11 nucleotides (range 0-30) compared to a median of 0 N-nucleotides in fetal tissues (liver range 0-2, bone marrow range 0-4), when the expressed sequences were compared to the reference genome. The junctional nucleotides contained many homopolymers ranging from dimers to hexamers, and trended towards higher guanosine content, consistent with TdT activity.
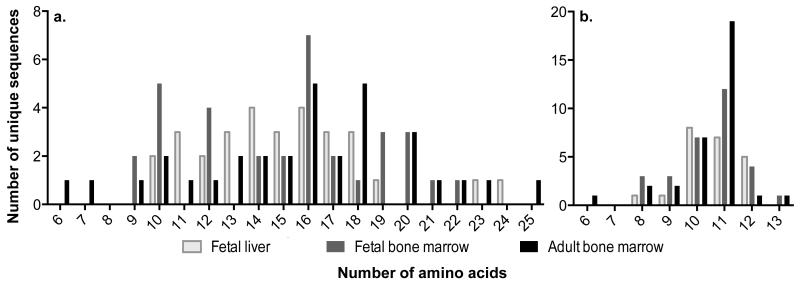
Although there was a difference in number of N-nucleotides, the median CDR3H length, which span the VDJ junctions was similar (p-values >0.3) in fetal (liver 15 codons, bone marrow 16 codons) and adult horse (bone marrow 16 codons) tissues (Fig. 6a). Similar results were found in the light chain: the median CDR3L length was 11 codons in all tissues (p-values >0.7) (Fig. 6b). The CDR3H and CDR3L lengths determined for the equine fetal liver and adult horse bone marrow were similar to those previously reported in the adult horse (Almagro et al. 2006; Sun et al. 2010; Tallmadge et al. 2013; Tallmadge et al. 2014).
Fig. 6. CDR3 lengths of equine fetal and adult horse immunoglobulins in primary lymphoid tissues.
Distribution of CDR3 amino acid lengths for the (a) heavy chain and (b) lambda light chain in the fetal liver (light gray), fetal bone marrow (dark gray), and adult horse bone marrow (black). No statistical differences were measured between the median CDR3 nucleotide lengths for fetal liver, fetal bone marrow, and adult horse bone marrow, all fetal and adult horse, or any two way comparisons for the Ig heavy (p-values >0.3) or lambda light (p-values >0.7) chains.
In order to further understand the similar CDR3 lengths despite a greater number of N-nucleotide additions in adult horse tissues, the lengths of IGHD segments and deletions off the 3′ end of the IGHV or IGLV and 5′ end of the IGHJ or IGLJ gene segments were examined. The median IGHD segment nucleotide lengths annotated in expressed sequences, subjected to exonuclease activity were significantly longer (p<0.005) in fetal liver (14 nucleotides, range 9-26) and fetal bone marrow (20, range 7-33) than those in the adult horse bone marrow (10, range 4-26). There was also significantly less (p<0.001) nucleotides removed from the 5′ end of the IGHJ gene segments of fetal sequences compared to the adult horse bone marrow, with no difference, however in the number of nucleotides deleted from the 3′ end of the IGHV segments (Supplemental Table 4). There were significantly less nucleotide removed from IGLV (p < 0.0004) and IGLJ (p < 0.0007) segments in fetal IGL sequences (Supplemental Table 4).
Discussion
We report herein hematopoietic activity in the liver and bone marrow of the equine fetus as early as 100DG. The mRNA expression of genes that represent hematopoiesis of lymphoid, myeloid, and erythroid lineages (Fig. 1) was observed in whole tissues and isolated cells from the fetal liver. Immunohistochemical (Fig. 2), flow cytometric (Table 1), and cytologic (Fig. 3) results of fetal liver tissues or isolated cells corroborate with active hematopoiesis, with predominant erythropoiesis. T and B cells were also observed at this developmental stage. T cells were mostly seen as isolated or small groups of cells, suggesting that they were likely circulating through the liver, as their primary development happens in the fetal thymus. B cells were seen isolated but more often in small clusters, suggesting B cell developmental niches. Although CD2 is considered typically a T cell marker, this molecule is also expressed in B cells, and tissues from one fetus displayed CD2+ cells in small clusters, similarly to the distribution of B cells. Sinkora et al. (1998) showed that, in the pig, CD2 is expressed in B cells during early development, and the number of CD2+ B cell decreases with age and exposure to microflora. It is possible that, like the pig, equine B cells express CD2 in fetal life, and further characterization of this B cell population is being pursued. Also similar to previous studies, a relatively large population of CD8+ cells was detected in the fetal liver with IHC. Many of the CD8+ cells observed in this study had morphologic characteristic of myeloid cells, which are known to express the CD8 molecule in other species, including humans (monocytes) (Gibbings et al. 2007) and rats (monocytes and macrophages), (Hirji et al. 1997; Lin et al. 2000) and further investigation is warranted. The presence of CD34+ cells supports the potential for hematopoiesis, and reveals the possibility for isolation and application of liver-derived HSC in regenerative and developmental studies. The percentage of CD34+ HSCs was assessed on frozen stored and thawed cells when an antibody with cross-reactivity to the equine antigen became available. Accordingly, the percentage of HSCs in the fetal liver may actually be greater than measured in this study if the equine HSCs are more sensitive to mortality than other leukocytes upon freezing.
While our results support the first part of our hypothesis that hematopoiesis occurs in the liver in early gestation with the generation of B cells, we were pleased to find that the Ig diversity was not as limited as expected, and no differential bias in IGHV segment use or CDR3 length between fetal and adult horse sequences were detected. Previously, we described no differences in heavy chain variable gene segments in secondary lymphoid tissue of the fetus and horse (Tallmadge et al. 2013). In this study, we show no differences in the primary lymphoid tissue IGH repertoire; the preference for IGHV2S3, IGHD18S1, and IGHJ1S5 to construct the heavy chain originates during fetal development, and is maintained in adulthood. In contrast, in this study we observed a bias for IGLV4-66 and IGLJ5 in fetal life but IGLV8-128 and IGLJ7 in the adult horse for the lambda light chain. Biases in V(D)J segment utilization are reported in fetal life in many species (Butler et al. 2006; Gontier et al. 2005; Jiang et al. 2011; Koti et al. 2010; Pascual et al. 1993), and have been proposed to be due to a variety of factors such as genomic location and accessibility (Souto-Carneiro et al. 2005; Yancopoulos et al. 1984; Zemlin et al. 2001), segment length and resulting CDR3 size (Shiokawa et al. 1999), and response to environmental conditions (Butler et al. 2000). However, the equine fetus uses broader range of IGLV segments, with the dominant IGLV segment present in 25% of fetal sequences but in 57% of adult horse lambda chain sequences, suggesting that none of the above mechanisms regulate Ig variable gene segment use in the horse. There were also a greater number of different IGLV-IGLJ combinations observed in the fetus (16 liver, 18 bone marrow) than the adult (14). Decreased restriction during fetal life also occurs in sheep, in which there is no bias for IGHJ selection in fetal or neonatal stages but IGHJ1 is most frequently used in the adult (Gontier et al. 2005). In contrast, humans (Lee et al. 2000) and swine (Butler et al. 2013; Wertz et al. 2013) have restricted IGLV utilization in their pre-immune Ig repertoires. It remains unknown what governs the shift to a predominance of IGLV8-128 and IGLJ7 in the adult horse. The same biases have been observed in horses of different breeds and geographic location (Hara et al. 2012; Sun et al. 2010; Tallmadge et al. 2014). In the fetus, the bias for IGLV4-66 and IGLJ5 may reflect the predominance of B1-like cells in this life stage, as B1 cells in other species preferentially use different V(D)J segments than B2 cells (Berland and Wortis 2002). IGLJ5, IGLJ6, and IGLJ7 have identical nucleotide sequences and can only be distinguished by the associated IGLC segment (Sun et al. 2010); therefore, the bias of IGLJ7 by the adult horse observed in this study is likely biologically insignificant from that of IGLJ5 in the fetus. The diversity of the pre-immune antibody repertoire in the primary lymphoid tissues of the equine fetus was assessed by analyzing 30-33 unique and productive immunoglobulin sequences donated by 3 different fetuses and adults (except for fetal liver, where IGL n=22 as discussed previously). Albeit relatively limited, the sequences were sufficient to demonstrate Ig combinatorial diversity in B cells produced during the fetal developmental period. The use of 3 different fetuses of distinct breeding parents accounts for individual variations; however, expansion of data with sequencing and bioinformatic technologies are being pursued to confirm and reveal further similarities and differences in the equine fetal and adult antibody repertoire.
CDR3 length influences the folding pattern, shape, and positioning of key amino acids in the CDR3 loop and, consequently, antigen specificity of the antibody (Barrios et al. 2004; Miqueu et al. 2007). The length of the CDR3 region is the result of both combinatorial and junctional diversity. The lengths of the V(D)J segments, bases removed from these segments by exonucleases, and N- and P-nucleotides added at the junction all contribute to CDR3 length (Desiderio et al. 1984; Lafaille et al. 1989; Sanz 1991). By 100DG, there were significantly less N-nucleotides at the IGHV-IGHD, IGHD-IGHJ, and IGLV-IGLJ junctions. P-nucleotides could not be confidently identified, as there were few full-length exons annotated; however, it was suspected that 17% fetal and 6% adult horse heavy chain sequences contained P-nucleotides (Supplemental Table 2). Despite the difference in N-nucleotide additions, equine fetal liver CDR3 lengths are similar to that of the adult horse as previously shown in secondary lymphoid tissue (Tallmadge et al. 2013). The length of the genomic V(D)J segments used could not fully explain this discrepancy; alternatively, the similar CDR3 lengths could be due to greater exonuclease activity in the adult horse or a greater number of mutations preventing the annotation of the actual 5′ and 3′ ends of the segments. Swine also have similar CDR3H lengths in fetal and adult life with early expression of TdT, and the longer IGHD segments used in fetal life are trimmed to a greater extent by exonucleases (Butler et al. 2000; Sinkora et al. 2002). In contrast to horses (Tallmadge et al. 2013; Tallmadge et al. 2014) and pigs (Butler et al. 2000), CDR3H lengths in humans (Delassus et al. 1998; Shiokawa et al. 1999) and mice (Bangs et al. 1991) increase with development. Longer CDR3s increase the potential nucleotide diversity, and restricting this length is hypothesized to be one of the mechanisms controlling the range of antigen binding sites during ontogeny (Shiokawa et al. 1999; Zemlin et al. 2001).
Pairwise nucleotide identity was higher in fetal life for the heavy chain, while no difference was observed at the lambda light chain locus, likely due to the wider variety of IGLV segments belonging to different subfamilies, composing the majority (~350bp) of the variable region (~400bp). Heavy and lambda light chain sequences expressed in fetal tissues have a higher percentage of identical nucleotides with the reference genome sequence than adult sequences; this difference is expected, as adult Ig molecules undergo somatic hypermutation and affinity maturation in response to antigenic stimulation in the periphery. Fetal tissues contain essentially naïve B cells, while those of the adult are colonized by newly generated naïve B cells in addition to antigenic experienced plasma cells (Nagasawa 2006).
Though at a lesser degree than that in the adult horse, fetal sequences did contain a small number of mutations identified by comparing the expressed sequences to that of the reference genome. Different allotypes have been described at the equine lambda locus, and nucleotide differences identified herein from the reference genomic sequence could be the result of allelic variation, sequence variation between different gene segments, and mutations (Hara et al. 2012; Sun et al. 2010; Tallmadge et al. 2014). Expressed IGL sequences in the fetus and the reference genomic sequences differed in nucleotide identity by as much as 9% (Supplemental Table 3); this difference is greater than the 4% of nucleotide differences between some IGL gene alleles observed (Hara et al. 2012; Tallmadge et al. 2014), likely reflecting sequence variation between IGL gene segments and mutations. Cattle fetus immunoglobulin genes also present mutations and undergo a small degree of somatic hypermutation in the absence of external antigen (Koti et al. 2010). Somatic hypermutation greatly contribute to antibody diversity, as HC1+/0 IgH−/− Igκ−/− mice express only one IGHV gene, and can respond to a number of different antigens with high affinity using junctional diversity and somatic hypermutation alone (Xu and Davis 2000). Therefore, even the minimal mutations in the V(D)J sequences generated in the presumed sterile environment of the womb may significantly contribute to the diversity of the equine Ig repertoire during fetal life.
Our study suggests that the B cells produced in the liver and bone marrow of the equine fetus generate a wide repertoire of pre-immune Igs for protection The B cells that develop in the primary lymphoid tissues during gestation have already elements of combinatorial, junctional, and sequence diversity in the Ig repertoire observed in the adult horse, with similar heavy chain VDJ segments and CDR3 lengths. By 100DG, we observed minor combinatorial diversity differences between the fetal liver and fetal bone marrow tissues, which suggests common regulatory mechanisms occurring at the two hematopoietic sites at this stage of development. A smaller degree of sequence diversity was also detected in the fetal primary lymphoid tissues, presumably reflecting the assumed sterile environment of the womb. The use of a greater number of different lambda variable gene segments in fetal life may provide the neonate an opportunity to respond to a wider range of antigens at birth.
Supplementary Material
Acknowledgements
We would like to thank Steven C. Miller and Mary Beth Matychak for their technical support. This research was partially funded by the NIH Director’s New Innovator Award 1 DP2 OD007216.
Abbreviations
- Ig
immunoglobulin
- DG
days of gestation
- HSC
hematopoietic stem cell
- mRNA
messenger RNA
- RT-PCR
reverse transcription polymerase chain reaction
- CD
cluster of differentiation
- CDR
complementary determining region
- AGM
aorta-gonad-mesonephros
- YS
yolk sac
- IGHV
Ig heavy chain variable gene
- IGHD
Ig heavy chain diversity gene
- IGHJ
Ig heavy chain joining gene
- IGLV
Ig lambda light chain variable gene
- IGLJ
Ig lambda light chain joining gene
- IGLC
Ig lambda light chain constant gene
- IGKV
Ig kappa light chain variable gene
- IGKJ
Ig kappa light chain joining gene
- IGKC
Ig kappa light chain constant gene
- TdT
terminal deoxynucleotidyl transferase
- N-nucleotide
non-template nucleotide
- P-nucleotide
palindromic nucleotide
- PBS
phosphate buffered solution
- cDNA
complementary DNA
- dNTPs
the four deoxynucleotide triphosphates
- M-MuLV
Moloney Murine Leukemia Virus
- CLP
common lymphoid progenitor
- TBS
tris buffered saline
- RACE
rapid amplification of cDNA ends
- MHC
major histocompatibility complex
- AEC
3-amino-9-ethylcarbazole
- FITC
fluorescein isothiocyanate
- LB broth
Luria-Bertani broth
References
- Almagro JC, Martinez L, Smith SL, Alagon A, Estevez J, Paniagua J. Analysis of the horse V(H) repertoire and comparison with the human IGHV germline genes, and sheep, cattle and pig V(H) sequences. Mol Immunol. 2006;43:1836–1845. doi: 10.1016/j.molimm.2005.10.017. doi: 10.1016/j.molimm.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Bangs LA, Sanz IE, Teale JM. Comparison of D, JH, and junctional diversity in the fetal, adult, and aged B cell repertoires. J Immunol. 1991;146:1996–2004. [PubMed] [Google Scholar]
- Barrios Y, Jirholt P, Ohlin M. Length of the antibody heavy chain complementarity determining region 3 as a specificity-determining factor. J Mol Recognit. 2004;17:332–338. doi: 10.1002/jmr.679. doi: 10.1002/jmr.679. [DOI] [PubMed] [Google Scholar]
- Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–997. doi: 10.1016/0092-8674(90)90502-6. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833. doi: 10.1146/annurev.immunol.20.100301.064833. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Sun X. Antibody repertoire development in fetal and neonatal piglets. XIV. Highly restricted IGKV gene usage parallels the pattern seen with IGLV and IGHV. Mol Immunol. 2013;55:329–336. doi: 10.1016/j.molimm.2013.03.011. doi: 10.1016/j.molimm.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Butler JE, Sun X, Wertz N, Lager KM, Chaloner K, Urban J, Jr, Francis DL, Nara PL, Tobin GJ. Antibody repertoire development in fetal and neonatal piglets XXI. Usage of most VH genes remains constant during fetal and postnatal development. Mol Immunol. 2011;49:483–494. doi: 10.1016/j.molimm.2011.09.018. doi: 10.1016/j.molimm.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Butler JE, Sun J, Wertz N, Sinkora M. Antibody repertoire development in swine. Dev Comp Immunol. 2006;30:199–221. doi: 10.1016/j.dci.2005.06.025. doi: 10.1016/j.dci.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Butler JE, Weber P, Sinkora M, Sun J, Ford SJ, Christenson RK. Antibody repertoire development in fetal and neonatal piglets. II. Characterization of heavy chain complementarity-determining region 3 diversity in the developing fetus. J Immunol. 2000;165:6999–7010. doi: 10.4049/jimmunol.165.12.6999. doi: 10.4049/jimmunol.165.12.6999. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Das S, Nozawa M, Klein J, Nei M. Evolutionary dynamics of the immunoglobulin heavy chain variable region genes in vertebrates. Immunogenetics. 2008;60:47–55. doi: 10.1007/s00251-007-0270-2. doi: 10.1007/s00251-007-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delassus S, Darche S, Kourilsky P, Cumano A. Ontogeny of the heavy chain immunoglobulin repertoire in fetal liver and bone marrow. J Immunol. 1998;160:3274–3280. [PubMed] [Google Scholar]
- Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, Landau N, Alt FW, Baltimore D. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311:752–755. doi: 10.1038/311752a0. doi:10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Douglas RH, Ginther OJ. Effects of repeated daily injections of prostaglandin F2alpha on ovaries in mares. Prostaglandins. 1976;12:881–894. doi: 10.1016/0090-6980(76)90061-7. doi: 10.1016/0090-6980(76)90061-7. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.5. 2011 http://www.geneious.com.
- Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaminio MJ, Rush BR, Davis EG, Hennessy K, Shuman W, Wilkerson MJ. Characterization of peripheral blood and pulmonary leukocyte function in healthy foals. Vet Immunol Immunopathol. 2000;73:267–285. doi: 10.1016/s0165-2427(00)00149-5. doi: 10.1016/S0165-2427(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Ford JE, Home WA, Gibson DM. Light chain isotype regulation in the horse. Characterization of Ig kappa genes. J Immunol. 1994;153:1099–1111. [PubMed] [Google Scholar]
- Fry TJ, Mackall CL. Interleukin-7: from bench to clinic. Blood. 2002;99:3892–3904. doi: 10.1182/blood.v99.11.3892. doi: 10.1182/blood.V99.11.3892. [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Marcet-Palacios M, Sekar Y, Ng MC, Befus AD. CD8 alpha is expressed by human monocytes and enhances Fc gamma R-dependent responses. BMC Immunol. 2007;8:12. doi: 10.1186/1471-2172-8-12. doi: 10.1186/1471-2172-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontier E, Ayrault O, Godet I, Nau F, Ladeveze V. Developmental progression of immunoglobulin heavy chain diversity in sheep. Vet Immunol Immunopathol. 2005;103:31–51. doi: 10.1016/j.vetimm.2004.08.013. doi: 10.1016/j.vetimm.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Hara S, Diesterbeck US, Konig S, Czerny CP. Transcriptional analysis of equine lambda-light chains in the horse breeds Rhenish-German Coldblood and Hanoverian Warmblood. Vet Immunol Immunopathol. 2012;145:50–65. doi: 10.1016/j.vetimm.2011.10.006. doi: 10.1016/j.vetimm.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Hirji N, Lin TJ, Befus AD. A novel CD8 molecule expressed by alveolar and peritoneal macrophages stimulates nitric oxide production. J Immunol. 1997;158:1833–1840. [PubMed] [Google Scholar]
- Hood L, Gray WR, Sanders BG, Dreyer WJ. Light chain evolution. Cold Spring Harb Symp Quant Biol. 1967;32:133–146. doi: 10.1101/SQB.1967.032.01.021. [Google Scholar]
- Jiang N, Weinstein JA, Penland L, White RA, 3rd, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci USA. 2011;108:5348–5353. doi: 10.1073/pnas.1014277108. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. Int J Biochem Cell Biol. 2008;40:22–27. doi: 10.1016/j.biocel.2007.01.026. doi: 10.1016/j.biocel.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Kaushik AK, Kehrli ME, Jr, Kurtz A, Ng S, Koti M, Shojaei F, Saini SS. Somatic hypermutations and isotype restricted exceptionally long CDR3H contribute to antibody diversification in cattle. Vet Immunol Immunopathol. 2009;127:106–113. doi: 10.1016/j.vetimm.2008.09.024. doi: 10.1016/j.vetimm.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Kelus AS, Weiss S. Variant strain of rabbits lacking immunoglobulin kappa polypeptide chain. Nature. 1977;265:156–158. doi: 10.1038/265156a0. doi: 10.1038/265156a0. [DOI] [PubMed] [Google Scholar]
- Kent D, Copley M, Benz C, Dykstra B, Bowie M, Eaves C. Regulation of hematopoietic stem cells by the steel factor/KIT signaling pathway. Clin Cancer Res. 2008;14:1926–1930. doi: 10.1158/1078-0432.CCR-07-5134. doi: 10.1158/1078-0432.CCR-07-5134. [DOI] [PubMed] [Google Scholar]
- Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27:619–628. doi: 10.1200/JCO.2008.17.9812. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koti M, Kataeva G, Kaushik AK. Novel atypical nucleotide insertions specifically at VH-DH junction generate exceptionally long CDR3H in cattle antibodies. Mol Immunol. 2010;47:2119–2128. doi: 10.1016/j.molimm.2010.02.014. doi: 10.1016/j.molimm.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Kydd J, Antczak DF, Allen WR, Barbis D, Butcher G, Davis W, Duffus WP, Edington N, Grunig G, Holmes MA. Report of the First International Workshop on Equine Leucocyte Antigens, Cambridge, UK, July 1991. Vet Immunol Immunopathol. 1994;42:3–60. doi: 10.1016/0165-2427(94)90088-4. doi: 10.1016/0165-2427(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Lafaille JJ, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- Lee J, Monson NL, Lipsky PE. The V lambda J lambda repertoire in human fetal spleen: evidence for positive selection and extensive receptor editing. J Immunol. 2000;165:6322–6333. doi: 10.4049/jimmunol.165.11.6322. doi: 10.4049/jimmunol.165.11.6322. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. Nomenclature of the human immunoglobulin genes. Curr Protoc Immunol Appendix 1:Appendix 1P. 2001 doi: 10.1002/0471142735.ima01ps40. doi: 10.1002/0471142735.ima01ps40. [DOI] [PubMed] [Google Scholar]
- Lin TJ, Hirji N, Stenton GR, Gilchrist M, Grill BJ, Schreiber AD, Befus AD. Activation of macrophage CD8: pharmacological studies of TNF and IL-1 beta production. J Immunol. 2000;164:1783–1792. doi: 10.4049/jimmunol.164.4.1783. doi: 10.4049/jimmunol.164.4.1783. [DOI] [PubMed] [Google Scholar]
- Lunn DP, Holmes MA, Antczak DF, Agerwal N, Baker J, Bendali-Ahcene S, Blanchard-Channell M, Byrne KM, Cannizzo K, Davis W, Hamilton MJ, Hannant D, Kondo T, Kydd JH, Monier MC, Moore PF, O’Neil T, Schram BR, Sheoran A, Stott JL, Sugiura T, Vagnoni KE. Report of the Second Equine Leucocyte Antigen Workshop, Squaw valley, California, July 1995. Vet Immunol Immunopathol. 1998;62:101–143. doi: 10.1016/s0165-2427(97)00160-8. doi: 10.1016/S0165-2427(97)00160-8. [DOI] [PubMed] [Google Scholar]
- Martin BR, Larson KA. Immune response of equine fetus to coliphage T2. Am J Vet Res. 1973;34:1363–1364. [PubMed] [Google Scholar]
- McNiece IK, Langley KE, Zsebo KM. The role of recombinant stem cell factor in early B cell development. Synergistic interaction with IL-7. J Immunol. 1991;146:3785–3790. [PubMed] [Google Scholar]
- Miqueu P, Guillet M, Degauque N, Dore JC, Soulillou JP, Brouard S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol Immunol. 2007;44:1057–1064. doi: 10.1016/j.molimm.2006.06.026. doi: 10.1016/j.molimm.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Montecino-Rodriguez E, Dorshkind K. New perspectives in B-1 B cell development and function. Trends Immunol. 2006;27:428–433. doi: 10.1016/j.it.2006.07.005. doi: 10.1016/j.it.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Morgan DO, Bryans JT, Mock RE. Immunoglobulins produced by the antigenized equine fetus. J Reprod Fertil. 1975;(Suppl (23)):735–738. [PubMed] [Google Scholar]
- Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat Rev Immunol. 2006;6:107–116. doi: 10.1038/nri1780. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- Nielsen JS, McNagny KM. CD34 is a key regulator of hematopoietic stem cell trafficking to bone marrow and mast cell progenitor trafficking in the periphery. Microcirculation. 2009;16:487–496. doi: 10.1080/10739680902941737. doi: 10.1080/10739680902941737. [DOI] [PubMed] [Google Scholar]
- Parrish CR, Carmichael LE, Antczak DF. Antigenic relationships between canine parvovirus type 2, feline panleukopenia virus and mink enteritis virus using conventional antisera and monoclonal antibodies. Arch Virol. 1982;72:267–278. doi: 10.1007/BF01315223. doi: 10.1007/BF01315223. [DOI] [PubMed] [Google Scholar]
- Pascual V, Verkruyse L, Casey ML, Capra JD. Analysis of Ig H chain gene segment utilization in human fetal liver. Revisiting the “proximal utilization hypothesis”. J Immunol. 1993;151:4164–4172. [PubMed] [Google Scholar]
- Perryman LE, McGuire TC, Torbeck RL. Ontogeny of lymphocyte function in the equine fetus. Am J Vet Res. 1980;41:1197–1200. [PubMed] [Google Scholar]
- Pinheiro A, Lanning D, Alves PC, Mage RG, Knight KL, van der Loo W, Esteves PJ. Molecular bases of genetic diversity and evolution of the immunoglobulin heavy chain variable region (IGHV) gene locus in leporids. Immunogenetics. 2011;63:397–408. doi: 10.1007/s00251-011-0533-9. doi: 10.1007/s00251-011-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL. Identification of Pax5 target genes in early B cell differentiation. J Immunol. 2008;180:1719–1728. doi: 10.4049/jimmunol.180.3.1719. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Garcia C, Hein WR, Weill JC. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Dahan A, Anquez V, Weill JC. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. T cell lineage commitment: identity and renunciation. J Immunol. 2011;186:6649–6655. doi: 10.4049/jimmunol.1003703. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saini SS, Kaushik A. Extensive CDR3H length heterogeneity exists in bovine foetal VDJ rearrangements. Scand J Immunol. 2002;55:140–148. doi: 10.1046/j.1365-3083.2002.01028.x. doi: 10.1046/j.1365-3083.2002.01028.x. [DOI] [PubMed] [Google Scholar]
- Sanz I. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J Immunol. 1991;147:1720–1729. [PubMed] [Google Scholar]
- Shimanuki M, Sonoki T, Hosoi H, Watanuki J, Murata S, Kawakami K, Matsuoka H, Hanaoka N, Nakakuma H. Molecular cloning of IGlambda rearrangements using long-distance inverse PCR (LDI-PCR) Eur J Haematol. 2013;90:59–67. doi: 10.1111/ejh.12037. doi: 10.1111/ejh.12037. [DOI] [PubMed] [Google Scholar]
- Shiokawa S, Mortari F, Lima JO, Nunez C, Bertrand FE, 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW., Jr IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol. 1999;162:6060–6070. [PubMed] [Google Scholar]
- Sinkora J, Rehakova Z, Sinkora M, Cukrowska B, Tlaskalova-Hogenova H, Bianchi AT, De Geus B. Expression of CD2 on porcine B lymphocytes. Immunology. 1998;95:443–449. doi: 10.1046/j.1365-2567.1998.00621.x. doi: 10.1046/j.1365-2567.1998.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkora M, Sinkorova J, Butler JE. B cell development and VDJ rearrangement in the fetal pig. Vet Immunol Immunopathol. 2002;87:341–346. doi: 10.1016/s0165-2427(02)00062-4. doi: 10.1016/S0165-2427(02)00062-4. [DOI] [PubMed] [Google Scholar]
- Souto-Carneiro MM, Sims GP, Girschik H, Lee J, Lipsky PE. Developmental changes in the human heavy chain CDR3. J Immunol. 2005;175:7425–7436. doi: 10.4049/jimmunol.175.11.7425. doi: 10.4049/jimmunol.175.11.7425. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang C, Wang Y, Zhang T, Ren L, Hu X, Zhang R, Meng Q, Guo Y, Fei J, Li N, Zhao Y. A comprehensive analysis of germline and expressed immunoglobulin repertoire in the horse. Dev Comp Immunol. 2010;34:1009–1020. doi: 10.1016/j.dci.2010.05.003. doi: 10.1016/j.dci.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Tallmadge RL, Tseng CT, Felippe MJ. Diversity of immunoglobulin lambda light chain gene usage over developmental stages in the horse. Dev Comp Immunol. 2014;46:174–179. doi: 10.1016/j.dci.2014.04.001. doi: 10.1016/j.dci.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallmadge RL, Tseng CT, King RA, Felippe MJ. Developmental progression of equine immunoglobulin heavy chain variable region diversity. Dev Comp Immunol. 2013;41:33–43. doi: 10.1016/j.dci.2013.03.020. doi: 10.1016/j.dci.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallmadge RL, McLaughlin K, Secor E, Ruano D, Matychak MB, Flaminio MJ. Expression of essential B cell genes and immunoglobulin isotypes suggests active development and gene recombination during equine gestation. Dev Comp Immunol. 2009;33:1027–1038. doi: 10.1016/j.dci.2009.05.002. doi: 10.1016/j.dci.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Timens W, Kamps WA. Hemopoiesis in human fetal and embryonic liver. Microsc Res Tech. 1997;39:387–397. doi: 10.1002/(SICI)1097-0029(19971201)39:5<387::AID-JEMT1>3.0.CO;2-E. doi: 10.1002/(SICI)1097-0029(19971201)39:5<387::AID-JEMT1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61:800–830. doi: 10.1002/iub.226. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- Verma S, Aitken R. Somatic hypermutation leads to diversification of the heavy chain immunoglobulin repertoire in cattle. Vet Immunol Immunopathol. 2012;145:14–22. doi: 10.1016/j.vetimm.2011.10.001. doi: 10.1016/j.vetimm.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Wang H, Morse HC., 3rd IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res. 2009;43:109–117. doi: 10.1007/s12026-008-8055-8. doi: 10.1007/s12026-008-8055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz N, Vazquez J, Wells K, Sun J, Butler JE. Antibody repertoire development in fetal and neonatal piglets. XII. Three IGLV genes comprise 70% of the pre-immune repertoire and there is little junctional diversity. Mol Immunol. 2013;55:319–328. doi: 10.1016/j.molimm.2013.03.012. doi: 10.1016/j.molimm.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Wu TT, Kabat EA. An Analysis of the Sequences of the Variable Regions of Bence Jones Proteins and Myeloma Light Chains and their Implications for Antibody Complementarity. J. Exp. Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. doi: 10.1016/S1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- Yokota T, Huang J, Tavian M, Nagai Y, Hirose J, Zuniga-Pflucker JC, Peault B, Kincade PW. Tracing the first waves of lymphopoiesis in mice. Development. 2006;133:2041–2051. doi: 10.1242/dev.02349. doi: 10.1242/dev.02349. [DOI] [PubMed] [Google Scholar]
- Zemlin M, Bauer K, Hummel M, Pfeiffer S, Devers S, Zemlin C, Stein H, Versmold HT. The diversity of rearranged immunoglobulin heavy chain variable region genes in peripheral blood B cells of preterm infants is restricted by short third complementarity-determining regions but not by limited gene segment usage. Blood. 2001;97:1511–1513. doi: 10.1182/blood.v97.5.1511. doi: 10.1182/blood.V97.5.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.