Abstract
Pescadillo (PES1) and the upstream binding factor (UBF1) play a role in ribosome biogenesis, which regulates cell size, an important component of cell proliferation. We have investigated the effects of PES1 and UBF1 on the growth and differentiation of cell lines derived from 32D cells, an interleukin-3 (IL-3)-dependent murine myeloid cell line. Parental 32D cells and 32D IGF-IR cells (expressing increased levels of the type 1 insulin-like growth factor I [IGF-I] receptor [IGF-IR]) do not express insulin receptor substrate 1 (IRS-1) or IRS-2. 32D IGF-IR cells differentiate when the cells are shifted from IL-3 to IGF-I. Ectopic expression of IRS-1 inhibits differentiation and transforms 32D IGF-IR cells into a tumor-forming cell line. We found that PES1 and UBF1 increased cell size and/or altered the cell cycle distribution of 32D-derived cells but failed to make them IL-3 independent. PES1 and UBF1 also failed to inhibit the differentiation program initiated by the activation of the IGF-IR, which is blocked by IRS-1. 32D IGF-IR cells expressing PES1 or UBF1 differentiate into granulocytes like their parental cells. In contrast, PES1 and UBF1 can transform mouse embryo fibroblasts that have high levels of endogenous IRS-1 and are not prone to differentiation. Our results provide a model for one of the theories of myeloid leukemia, in which both a stimulus of proliferation and a block of differentiation are required for leukemia development.
Growth in the size of an individual cell is a fundamental growth process, as cell division requires growth in the size of the cell (13, 19). Cell size is essentially dependent on ribosome biogenesis (23). Ribosome biogenesis is controlled by the rate of rRNA synthesis (37), which is dependent on the activity of RNA polymerase I (10, 16, 28, 48). Thus, cell size is regulated by RNA polymerase I and the proteins that modulate its activity. Among the proteins which regulate the activity of RNA polymerase I is the upstream binding factor (UBF1) (16). While an increase in cell size is necessary for cell proliferation, cell division also requires the implementation of the cell cycle program. An important question is how the two programs, increase in cell size and cell cycle progression, are coordinated. Recent reports have indicated that certain proteins are involved in both the cell cycle program and ribosome biogenesis (15, 21, 33, 41, 67). One of these proteins is pescadillo (also called PES1 and Yph1p), reported to be involved in DNA replication and ribosome biogenesis (12, 31).
In many hemopoietic cell lines, the induction of differentiation is preceded by a period of vigorous cell proliferation, and it has been suggested that this period of cell proliferation is necessary for differentiation to occur (65, 71). It seems that, in hemopoietic cells, certain growth factors send both proliferation and differentiation signals (38), with the latter eventually prevailing. Therefore, at least in certain cell types, a third component may be necessary for sustained cell proliferation—i.e., the extinction of a differentiation program. This hypothesis is not novel. For years, a number of investigators have proposed that a block in differentiation is a sine qua non for the development of malignancy, particularly in hemopoietic tumors (14, 24, 42, 43, 54, 60). Indeed, the use of retinoic acid for some forms of leukemia has been referred to as a differentiation therapy (6). This situation has been elegantly summarized by Gilliland and Tallman (14), who proposed two classes of mutations in acute leukemia: one class of mutations confers a proliferative advantage, while a second class of mutations impairs hemopoietic differentiation.
The respective roles of proliferation and differentiation in determining the fate of hemopoietic cells are well illustrated for 32D murine myeloid cells. 32D cells require interleukin-3 (IL-3) for growth and rapidly undergo apoptosis after the withdrawal of IL-3 (65, 74). Parental 32D cells have very low levels of the type 1 insulin-like growth factor I (IGF-I) receptor (IGF-IR) and do not express insulin receptor substrate (IRS) 1 (IRS-1) or IRS-2 (64, 70, 73), the main docking protein of both the IGF-IR and the insulin receptor (72). 32D cells expressing moderate levels of a human IGF-IR (32D IGF-IR cells) survive in the absence of IL-3 and, with the addition of IGF-I, grow vigorously for about 48 h (64). After 48 to 72 h, 32D IGF-IR cells begin to differentiate along the granulocytic pathway (64). This action is not surprising, as IGF-I can induce differentiation in other hemopoietic cell lines besides the 32D cell line (1, 32). When 32D IGF-IR cells are stably transfected with a plasmid expressing IRS-1 (32D IGF-IR/IRS-1 cells), the cells no longer differentiate, grow indefinitely in the absence of IL-3, and even form tumors in mice (63). We interpret these results as indicating that in 32D cells, the activated IGF-IR, like other growth factor receptors of hemopoietic cells, sends signals for both proliferation and differentiation. When the cells express IRS-1, proliferation prevails, while in its absence, the cells differentiate. The antidifferentiation signal from IRS-1 is indirectly supported by the observation that cell types prone to differentiation often do not express IRS-1 or express very small amounts (25, 36, 53, 59, 70).
IRS-1 also plays a major role in the regulation of cell and body size. Drosophila has a receptor that partakes of both the IGF-IR and the IR and has homologues of the IRS proteins phosphatidylinositol 3-kinase, Akt, and S6K1. All of these homologues regulate cell and body size in Drosophila (4, 30, 35, 66). For instance, deletion of the Drosophila IRS homologue chico reduces fly weight by 65% in females and 55% in males. The reduction in body and organ size is due to a reduction in both cell number and cell size. Mice with a targeted disruption of the IRS-1 (45) or S6K1 (57) gene are smaller than their wild-type littermates. Similarly, 32D cells expressing IRS-1 are twice as large as 32D cells not expressing IRS-1, even when both cell types are growing exponentially (63). The recent reports that IRS-1 can translocate to the nucleus and bind UBF1 (58, 62) provide a molecular explanation for the effect of IRS-1 on cell and body size. During differentiation (which is inhibited by IRS-1), an inhibition of RNA polymerase I activity (10, 11, 28, 61) and a decrease in cell size (63) occur. In terminally differentiated cells, UBF1 is no longer detectable (11, 61), and the nucleolus (which is the site of rRNA synthesis) involutes (50).
As indicated above, PES1 is involved in both ribosome biogenesis and DNA replication, while UBF1 is one of the regulators of RNA polymerase I activity. Our hypothesis is that 32D-derived cells require three different signals from the activated IGF-IR for sustained cell proliferation: an increase in cell size, the activation of the cell cycle program, and the extinction of a differentiation program. To test this hypothesis, we examined whether PES1 and UBF1 can make 32D IGF-IR cells IL-3 independent, like IRS-1 does. We found that both PES1 and UBF1 cause an increase in cell size, that PES1 alters the cell cycle distribution, but that neither of them can inhibit the differentiation program and make 32D IGF-IR cells grow indefinitely in medium deprived of IL-3 and supplemented with IGF-1. The results were different for two cell lines of untransformed mouse embryo fibroblasts (MEFs) that express substantial levels of IRS-1 and are therefore refractory to differentiation. In these cells, both PES1 and UBF1 cause cellular transformation, as measured by the ability to form colonies in soft agar. We suggest that, with regard to IGF-IR signaling in 32D cells, extinction of the differentiation program is a requirement for sustained cell proliferation. This suggestion is in agreement with the hypothesis mentioned above that in certain forms of leukemia, both a stimulation of proliferation and a block of differentiation are necessary for tumor development. In this respect, our experiments simply provide a molecularly defined model for the respective roles of proliferation and differentiation in the growth of a myeloid cell line that can either differentiate or be transformed into a tumor-forming cell line (63).
MATERIALS AND METHODS
Plasmids.
pRcCMV FLAG UBF1, containing the mouse UBF1 cDNA with a 5′ FLAG tag, was a gift from I. Grummt, German Cancer Research Center, Heidelberg, Germany. It was described by Voit et al. (67). 5′ FLAG UBF1 cDNA was excised with HindIII and XbaI, filled in with the Klenow enzyme, and cloned in the HindIII/HpaI sites of the MSCVpuro retroviral vector, generating the MSCVpuro FLAG/UBF1 retroviral vector. Selection was carried out with 1.5 μg of puromycin/ml. PES1 3′FLAG cDNA was generated by PCR from a mouse heart cDNA library (Clontech) with the following primers: forward primer, 5′-ACGTGGAGCTATGGGAGGTCTG-3′; and reverse primer, 5′-CTACTTGTCATCGTCGTCCTTGTAGTCAGGGACAACTGGAGCGCACAC-3′. PCR products were cloned in the Topo2.1 vector (Invitrogen) following the manufacturer's protocol. The PES1 cDNA sequence was the same as that in NCBI accession number AF289539. PES1 FLAG cDNA was excised from the Topo2.1 plasmid with BamHI/XhoI and ligated with BglII/XhoI in the MSCVpuro retroviral vector, generating the MSCVpuro PES1/FLAG retroviral vector.
Cell lines.
The cell lines used were 32D clone 3 (65), 32D IGF-IR (63), and 32D IGF-IR/IRS-1 (64). In one experiment, we also used 32D IRS-1 cells (74), which are parental 32D cells expressing IRS-1. 32D and 32D-derived cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies, Inc.), 10% WEHI cell-conditioned medium as a source of IL-3, and the antibiotic required for selection: 250 μg of G418 (Life Technologies)/ml or 250 μg of hygromycin (Calbiochem)/ml.
32D, 32D IGF-IR, 32D IGF-IR/IRS-1, and 32D IRS-1 cells were transduced (46) with the MSCVpuro FLAG/UBF1 and MSCVpuro PES1/FLAG retroviral vectors. Mixed populations were selected with 1.5 μg of puromycin/ml to generate the various cell lines described below.
In other experiments, we used as MEFs R12 and R508 cells. Both of these cell lines were derived from R− cells, which are MEFs originating from mouse embryos with a targeted disruption of the IGF-IR genes (56). R12 and R508 cells were generated by transfection with a plasmid expressing human IGF-IR. R12 cells express 3 × 103 IGF-IRs/cell, while R508 cells express 15 × 103 receptors/cell (52). Both of these cell lines do not respond to IGF-I with proliferation and do not form colonies in soft agar (49, 52). They were transduced with the same retroviral vectors and selected with the same procedures as 32D cells as described above.
Growth curves.
Cells exponentially growing in IL-3 were washed three times with Hanks balanced salt solution (HBSS) and seeded at a density of 5 × 104 cells/35-mm plate containing 2 ml of RPMI 1640 medium supplemented with 10% FBS, no IGF-I or IGF-I at 50 ng/ml (GIBCO BRL), or 10% WEHI cell-conditioned medium as the source of IL-3. Cells were counted by trypan blue exclusion (Life Technologies) at various times after IL-3 withdrawal.
Northern blotting.
Exponentially growing cells were washed three times with HBSS and seeded at 5 × 104 cells/ml in RPMI 1640 medium supplemented with 10% FBS, IGF-I at 50 ng/ml, 10% WEHI cell-conditioned medium, or granulocyte colony-stimulating factor at 25 ng/ml (BD Biosciences Pharmingen). At various times, cells were collected and total RNA was extracted with an RNeasy minikit (Qiagen). A total of 8 μg of total RNA for each sample was run on a 1% agarose formaldehyde gel, blotted onto a nylon membrane, and hybridized with 24p3 cDNA (46) or myeloperoxidase (MPO) cDNA (47).
Western blotting and immunoprecipitation.
Cells seeded under the same conditions as those used for Northern blotting were harvested at various times, washed with cold phosphate-buffered saline (PBS), and lysed in 50 mM HEPES (pH 7.5)-150 mM NaCl-1.5 mM MgCl2-1 mM EGTA-10% glycerol-1% NP-40-100 mM NaF-10 mM sodium pyrophosphate-0.2 mM sodium orthovanadate-1 mM phenylmethylsulfonyl fluoride-10 μg of aprotinin/ml. The rest of the procedure was the same as that described in detail in previous reports (39, 47). The following antibodies were used: anti-IRS-1 C-20 and anti-ID2 (Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-FLAG M2-peroxidase conjugate A-8592 and EZview red anti-FLAG M2 affinity gel F2426 (Sigma), anti-pescadillo rabbit polyclonal antibody (Bethyl Laboratories, Inc.), anti-glyceraldehyde-3-phosphate dehydrogenase (Research Diagnostics, Inc.), and anti-Grb2 (Transduction Laboratories).
FACS analysis.
Exponentially growing cells were washed with HBSS and seeded at 5 × 104 cells/ml in RPMI 1640 medium supplemented with 10% FBS, IGF-I at 50 ng/ml, or 10% WEHI cell-conditioned medium. After 24 h, the cells were washed with cold PBS and analyzed by fluorescence-activated cell sorting (FACS) for cell size (63). For cell cycle distribution, cells were fixed in 70% ethanol for 2 h on ice and then washed with PBS. Cells were suspended in propidium iodide-Triton X-100 staining solution with RNase A and analyzed after 30 min. For bromodeoxyuridine (BrdU) labeling, cells were incubated with BrdU for 1 h by using a BrdU flow kit (BD Biosciences Pharmingen) following the manufacturer's protocol. FACS analysis was carried out by using a Beckman Coulter XL four-color analyzer with an argon ion 488-nm laser (Beckman Coulter Corporation, Hialeah, Fla.).
Morphological analysis.
For analysis of differentiation, exponentially growing cells were collected, washed three times with HBSS, and seeded at 5 × 104 cells/ml in RPMI 1640 medium containing 10% FBS and 50 ng of IGF-I/ml. At various times, viable cells were counted by trypan blue exclusion (Life Technologies), and cytospin samples were used for morphological analysis as described by Valentinis et al. (64).
Confocal microscopy.
Cells plated on glass coverslips were washed with PBS and fixed with 3.0% paraformaldehyde in PBS for 20 min at room temperature, followed by permeabilization with 0.2% Triton X-100 in PBS for 2 min at room temperature. Coverslips were washed with PBS and blocked in 10% normal donkey serum (sc-2044; Santa Cruz Biotechnology) in PBS for 20 min at room temperature. Coverslips were washed with PBS and incubated for 1 h at room temperature with anti-FLAG-fluorescein isothiocyanate-conjugated F4049 (Sigma) antibody. After being washed with PBS, coverslips were digested with RNase A (1 mg/ml) for 30 min and then stained with propidium iodide (2.5 μg/ml; P-3566; Molecular Probes) for 5 min. Finally, coverslips were washed with PBS three times and mounted on glass slides with Vectashield mounting medium (H-1000; Vector Laboratories Inc.). Fluorescent images were collected by using a Zeiss Axiovert 100 confocal microscope with a Zeiss ×40 objective.
Statistical analysis.
Statistical significance (P < 0.05) between two measurements and among multiple groups was determined with the two-tailed Student t test analysis of variance and with the Bonferroni method, respectively (44).
RESULTS
Expression of exogenous PES1 and UBF1 in 32D-derived cells.
The original cell lines were described previously: parental 32D cells (65), 32D IGF-IR cells (44, 64), 32D IRS-1 cells (74), and 32D IGF-IR/IRS 1 cells (63). These four cell lines were infected with retroviruses expressing either UBF1 or PES1 to obtain stable mixed populations overexpressing these proteins in the four original cell lines. The expression of PES1 or UBF1 in infected cells is shown in Fig. 1, where the expression of the proteins was detected by Western blotting with an antibody to FLAG. The FLAG-tagged proteins were expressed vigorously in all four cell lines, but they were not detectable in uninfected parental 32D cells. The levels of expression of both PES1 and UBF1 seemed to be lower in infected parental 32D cells. We did not pursue this difference, since it is not relevant to the results presented below. The lower band in Fig. 1 is a nonspecific band (FLAG antibodies, in our experience, are better for immunoprecipitation than for Western blotting). After selection was completed, 99.9% of the cells were found to be PES1 positive (see below).
FIG. 1.
Expression of PES1 and UBF1 in 32D-derived cells. 32D, 32D IGF-IR, 32D IGF-IR/IRS-1, and 32D IRS-1 cells were infected with retroviruses expressing either PES1 or UBF1 as described in Materials and Methods. Mixed populations were collected, lysates were made, and Western blots were developed with an antibody to the FLAG epitope. Parental 32D cells were used as negative controls for the FLAG antibody. The positions of ectopic PES1 (A) and UBF1 (B) are indicated by arrows.
Growth of 32D-derived cells in IGF-1.
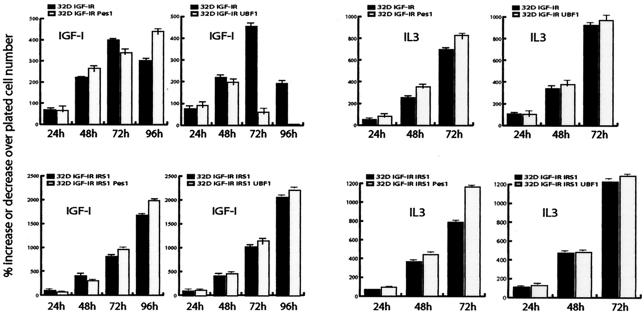
We first determined the growth of these cell lines under different conditions. As usual, all 32D cell lines died within 24 h after IL-3 withdrawal in medium not supplemented with IGF-I (data not shown). The growth of six cell lines is shown in Fig. 2. In medium supplemented with IGF-I, 32D IGF-IR cells grew vigorously for 48 h and then slowed down, reaching a maximum of a fourfold increase in number (44, 64). The expression of PES1 in 32D IGF-IR cells had no noticeable effect on their growth. 32D IGF-IR cells expressing PES1 grew exponentially for the first 48 to 72 h but then stopped growing, like their parental cell line. The expression of UBF1 in these cells actually had a negative effect, as the cell number decreased more rapidly than in parental 32D IGF-IR cells. All three cell lines grew exponentially for the first 72 h, after which they stopped growing (Fig. 2).
FIG. 2.
Growth curves for 32D-derived cell lines. The growth of the cell lines in either IGF-I (50 ng/ml) or IL-3 was determined. Open bars indicate parental cells, either 32D IGF-IR cells (upper panels) or 32D IGF-IR/IRS-1 cells (lower panels). The number of cells was counted at 24, 48, and 96 h after shifting to IGF-I or replating in IL-3. The numbers on the ordinate represent the percent increase over the number of plated cells (e.g., 300 indicates a threefold increase). Note the different ordinate scales for 32D IGF-IR and 32D IGF-IR/IRS-1 cells. We omitted an analysis of parental 32D cells, which grow very well in IL-3 and die quickly after a shift to IGF-I. Error bars indicate standard deviations.
32D IGF-IR/IRS-1 cells grow continuously in the absence of IL-3, if supplemented with IGF-I, and actually form tumors in mice (63). The expression of either UBF1 or PES1 in these cells made them grow just as well, if not a little better (Fig. 2). At 96 h after a shift to IGF-I, the cell number had increased by about 20-fold. With UBF1, we could not detect any significant increase in growth with respect to that of 32D IGF-IR/IRS-1 cells, but with PES1, we could detect a small but reproducible increase in the number of cells—between 10 and 15%. This modest effect of PES1 but not of UBF1 on the number of 32D IGF-IR/IRS-1 cells was also detectable in cells growing in IL-3 (Fig. 2).
Parental 32D cells died even in the presence of IGF-I, as expected, and the expression in these cells of either UBF1 or PES1 had very little effect on their survival. At the end of 24 h, a few more of these cells than of parental 32D cells were alive, but all cells had died by 48 h in all three cell lines (data not shown). We also infected with the same retroviruses 32D IRS-1 cells, which do not survive the shift from IL-3 to IGF-I (74). Neither PES1 not UBF1 protected these cells from apoptosis (data not shown). These experiments indicate that, unlike IRS-1, neither PES1 nor UBF1 can sustain the proliferation of 32D IGF-IR cells when the cells are shifted from IL-3 to IGF-I. PES1 has a modest positive effect on the growth of 32D IGF-IR/IRS-1 cells.
We therefore set out to investigate the mechanism(s) behind the failure of PES1 and UBF1 to sustain the growth of 32D IGF-IR cells.
Cell cycle distribution (DNA amounts).
Because PES1 is known to affect DNA synthesis, besides ribosome biogenesis (12, 26, 31), and also to slightly increase the growth of 32D IGF-IR/IRS-1 cells (Fig. 2), we examined whether it would alter cell cycle distribution. UBF1 has not been reported to affect DNA synthesis, and we did not expect it to affect cell cycle distribution. To determine the cell cycle distributions of various cell lines by FACS analysis (see Methods and Materials), we examined them 24 h after a shift from IL-3 to IGF-1. At this time, all cell lines were growing exponentially and at similar rates (Fig. 2), so that FACS analysis provides the cell cycle distributions of cell populations in which close to 100% of the cells are dividing (44). 32D IGF-IR cells and 32D IGF-IR/IRS-1 cells, whether expressing UBF1 or not, had the usual cell cycle distributions, with about 45% of cells in G1 phase, 35% in S phase, and 20% in G2 phase. Dramatically different results were obtained with cells expressing PES1: the percentage of cells in S phase was only 18%, while there was an accumulation of cells in the G2 peak (4N DNA) to as much as 47% (Fig. 3A). This was true whether PES1 was expressed in 32D IGF-IR or 32D IGF-IR/IRS-1 cells (Fig. 3A). This experiment was repeated with four different samples at 24 and 48 h, with similar results. This anomalous distribution seemed to be dependent on IGF-1 for two reasons. First, 32D cells expressing PES1 died even in the presence of IGF-1, and second, the abnormal S phase was hardly detectable in the same cells in the presence of IL-3. There was a slight tendency for an increase in the number of cells in the G2 peak even in the presence of IL-3 (data not shown), but this increase probably would have passed unnoticed without the dramatic results obtained when the cells were grown in IGF-1.
FIG. 3.
Cell cycle analysis of 32D-derived cells. 32D IGF-IR and 32D IGF-IR/IRS-1 cells and the same cell lines expressing exogenous PES1 or UBF1 were grown in IGF-1 (24 h after a shift from IL-3). (A) FACS analysis of six cell lines for cell cycle distribution. Under these conditions, all cell lines grew exponentially. In PES1-expressing cells, there was a marked increase in the fraction of cells in the G2 peak and a decrease in the fraction of cells with an S-phase amount of DNA (see the text). These experiments were repeated four times up to 6 days after a shift from IL-3 to IGF-I, with similar results (the differences in IL-3 were more modest). (B) Percentage of cells labeled by a 1-h exposure to BrdU. Error bars indicate standard deviations; values at right indicate means and standard deviations. Statistically significant differences are marked with an asterisk (comparison between each parental cell line and its PES1- and UBF1-expressing counterparts by a t test). All cell lines expressing PES1 showed a decrease in the percentage of BrdU-labeled cells.
The prominent G2 peak was present in cells expressing PES1 at various times after a shift to IGF-I, up to 6 days, the last time at which we monitored the cell cycle distribution (data not shown). However, at this point, the cells (with the exception of cells expressing IRS-1) had stopped growing, so that at this point a discussion of cell cycle phases would be meaningless. On day 6, the PES1-expressing cells were differentiated into granulocytes (see below). In reality, what we were measuring on day 6 was the DNA content of cells. However, during exponential growth, FACS analysis provides the cell cycle distributions of exponentially growing cells. This point is addressed again in the Discussion.
To confirm the decrease in the number of exponentially growing 32D-derived cells expressing PES1 in S phase, we labeled the cells with BrdU 24 h after a shift from IL-3 to IGF-I. The cells were labeled for 1 h, so that the fraction of labeled cells would include those that were synthesizing DNA when BrdU was added and those that entered S phase during the 1-h labeling. The results are summarized in Fig. 3B. UBF1 expression had little or no effect on the percentage of cells labeled by BrdU, but PES1 expression markedly decreased it. This finding was especially evident in 32D IGF-IR cells, where the expression of PES1 decreased the percentage of cells labeled by BrdU from 58 to 32%. At this time after a shift to IGF-1, all cell lines, except for the parental 32D cell line, are growing exponentially with about the same doubling times (44; see below). The decrease in the percentage of cells in S phase therefore is compatible with a shortening of S phase and a lengthening of G2 phase.
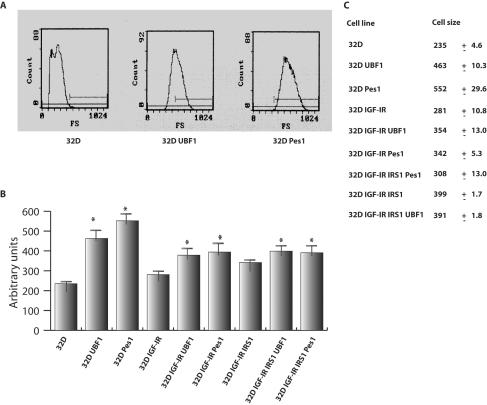
Cell size.
We next examined whether the overexpression of PES1 and UBF1 would alter cell size. Both UBF1 (16) and PES1 (12, 31) play an important role in ribosome biogenesis, which determines cell size (16, 23, 37). The expression of UBF1 in cardiomyocytes increases cell size (17). IRS-1, which binds to UBF1 (58, 62), also increases cell size, both in cells in cultures (63) and in animals (4, 45). Since parental 32D cells do not survive in the absence of IL-3, these experiments were carried out with cells growing in IL-3. The results of a representative experiment are shown in Fig. 4, where cell size was determined by FACS analysis. Both UBF1 and PES1 increased cell size, about as well as if not better than IRS-1. The increase was most dramatic in parental 32D cells, where both UBF1 and PES1 doubled the mean size of cells. The increase was less dramatic in 32D IGF-IR and 32D IGF-IR/IRS-1 cells, although it was significant and reproducible. In the last two cell lines, UBF1 and PES1 increased cell size by about 20 to 25%. Additional data on cell size are given below.
FIG. 4.
Cell size of 32D-derived cells expressing UBF1 or PES1. All cell lines were grown in IL-3. (A) Representative data from FACS analysis for cell size (angle scattering) only for 32D cells, either parental or expressing either UBF1 or PES1. (B) Similar analysis for other 32D-derived cells. Cell size was expressed in arbitrary units; error bars indicate standard deviations. The asterisks indicate significant differences (P < 0.05) between the parental cell lines and the cell lines expressing ectopic UBF1 or PES1, as determined by a t test. (C) Actual means and standard deviations for cell size. Overexpression of UBF1 and PES1 increased cell size for all three cell lines, although the increase was more dramatic for parental 32D cells (see the text). These experiments were done with cells in IL-3, but the same results were obtained with cells in IGF-I, at either 2 or 6 days after a shift from IL-3 to IGF-I.
PES1 and UBF1 fail to inhibit the differentiation of 32D IGF-IR cells.
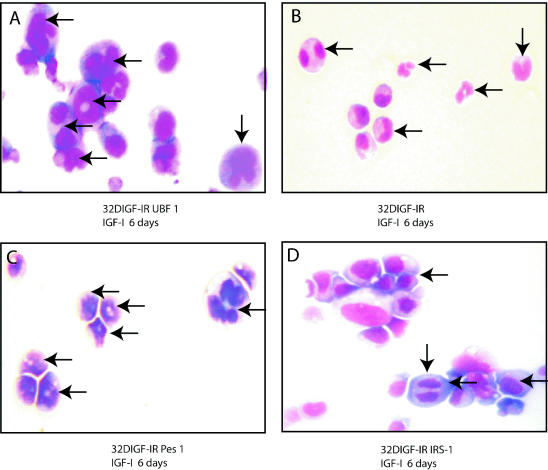
32D IGF-IR cells, after a shift from IL-3 to IGF-I, grow for at least 48 h and then differentiate into granulocytes. In contrast, 32D IGF-IR/IRS-1 cells keep growing and do not show any sign of differentiation (63, 64). We tested the hypothesis that 32D IGF-IR cells expressing PES1 or UBF1 stop growing because they also differentiate, like the parental cells. This hypothesis was definitively confirmed by morphological analysis (Fig. 5). When the parental and derived 32D IGF-IR cell lines were examined 6 days after a shift to IGF-I, it was clear that the cells were differentiating along the granulocytic pathway. The most obviously differentiated cells are visible in Fig. 5A to C, but most cells were in one or another phase of differentiation. 32D IGF-IR/IRS-1 cells (Fig. 5D) were the only cells not showing features of differentiation; the nuclei were round, and even one mitosis event was visible. Figure 5 also confirms the dramatic increase in cell size caused by either PES1 or UBF1. Despite the fact that the cells were differentiated, they were obviously larger than parental 32D IGF-IR cells (Fig. 5B). FACS analysis of these same cells at 6 days after the shift to IGF-I confirmed the marked increase in cell size (data not shown). 32D IGF-IR/IRS-1 cells were also larger than 32D IGF-IR cells, as previously reported (63). These results indicate that PES1 and UBF1 cannot inhibit the differentiation program initiated by IGF-IR in 32D cells, although they can have some effects on DNA replication and cell size.
FIG. 5.
Differentiation of 32D-derived cells into granulocytes. The cells examined were 32D IGF-IR cells, 32D IGF-IR cells expressing PES1, 32D IGF-IR cells expressing UBF1, and 32D IGF-IR/IRS-1 cells. The cells were stained with Giemsa stain 6 days after a shift from IL-3 to IGF-I. All images were taken at the same magnification, ×40. The arrows in panels A, B, and C indicate differentiating cells (bilobed, irregular nuclei and some well-differentiated granulocytes). The 32D IGF-IR/IRS-1 cells in panel D have round nuclei, and one of them (vertical arrow) is actually in mitosis. Note the difference in size between parental 32D IGF-IR cells (B) and the same cells expressing PES1 (C), UBF1 (A), or IRS-1 (D).
Although granulocytes become detectable only after 4 to 5 days, markers of differentiation appear within the first 24 h (47, 64). Since 32D IGF-IR cells expressing either PES1 or UBF1 stop growing after 48 to 72 h in IGF-I, we wanted to know whether differentiation markers would also appear early in 32D IGF-IR cells expressing PES1 or UBF1. As markers of differentiation, we chose MPO mRNA, 24p3 mRNA (55), and ID2 protein. IRS-1 has been shown to inhibit the expression of MPO and 24p3 mRNAs (46, 47, 63) and to induce ID2 protein expression (2, 47). Figure 6 shows that in 32D IGF-IR cells, a shift to IGF-I causes a sharp increase in the expression of MPO and 24p3 mRNAs and no increase in the expression of ID2 protein. The expression of IRS-1 in these cells, as already reported, blocks differentiation, as evidenced by the induction of ID2 protein expression and the repression of MPO and 24p3 mRNAs. The expression of either PES1 or UBF1 gives the same results as in parental 32D IGF-IR cells (Fig. 6). PES1 and UBF1 also fail to induce the expression of ID2 protein. These results confirm that 32D IGF-IR cells expressing either PES1 or UBF1 promptly initiate a differentiation program in IGF-1 -supplemented medium.
FIG. 6.
Expression of MPO and 24p3 mRNAs and of ID2 protein in 32D-derived cells. (Upper panel) Northern blots of mRNAs for two markers of differentiation, 24p3 and MPO. Times, cell lines, and treatments are indicated above the lanes. Only IRS-1 inhibited the appearance of these markers in 32D IGF-IR cells. The bottom section gives the mRNA amount in each lane. (Lower panels) Western blots showing the expression of the ID2 protein, which was markedly increased in 32D IGF-IR cells, in which differentiation was inhibited by IRS-1 (2, 47). Only IRS-1 was capable of inducing ID2 expression. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Interaction between PES1 and IRS-1.
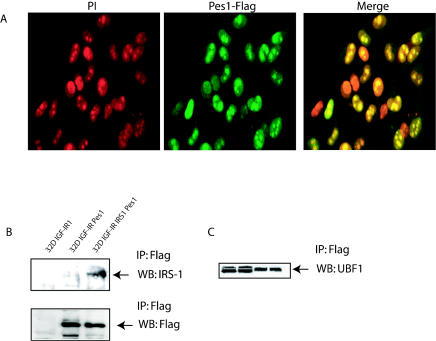
IRS-1 can localize to nucleoli (58, 62), where it interacts with UBF1, an exclusively nucleolar protein (61, 68). Since PES1 has been reported to localize to nucleoli (31), we examined whether there was also an interaction between PES1 and IRS-1. First, we confirmed by confocal microscopy that PES1 could localize to nucleoli. Cells infected with a retrovirus expressing PES1 were stained with propidium iodide and an anti-FLAG antibody and examined earlier during selection. Figure7A shows that some of the cells expressed variable amounts of ectopic PES1. Among the cells expressing higher levels of FLAG-tagged PES1, it was clear that the protein localized to nuclei and to nucleoli.
We next studied the PES1-IRS-1 interaction by coimmunoprecipitation. Lysates were prepared and immunoprecipitated with an anti-FLAG antibody (which recognizes only exogenous PES1). Figure 7B shows that the anti-FLAG antibody immunoprecipitated IRS-1 in 32D IGF-IR/IRS-1 cells (the other two cell lines were used as controls not expressing IRS-1). The FLAG-tagged protein, though, was also visible in 32D IGF-IR cells expressing PES1 (Fig. 7B). We also studied the interaction between PES1 and UBF1. In lysates of cells expressing the FLAG-tagged PES1 construct, immunoprecipitation with an anti-FLAG antibody caused the appearance of UBF1 in the immunoprecipitate (Fig. 7C). Two negative controls were used. In one control experiment, lysates from cells not expressing the FLAG-tagged PES1 construct were immunoprecipitated with the anti-FLAG antibody. No IRS-1 was detected in the immunoprecipitate. In the second control experiment, we immunoprecipitated with the anti-FLAG antibody FLAG-tagged mutant IRS-1 (46) that does not localize to nuclei (deletion of the PHPTB domain). We could not detect endogenous PES1 in the immunoprecipitate (data not shown). It seems, therefore, that UBF1, IRS-1, and PES1 all can colocalize to nucleoli, where they may form a large complex cooperating in the activation of the ribosomal DNA promoter (16).
FIG. 7.
PES1 localizes to the nuclei and nucleoli of cells. (A) Confocal microscopy of cells stained with propidium iodide (PI) (red) and counterstained with an anti-FLAG antibody (green). The merged image shows the localization of PES1 (FLAG tagged) within the nuclei and nucleoli of infected cells. (B and C) Interactions among IRS-1, PES1, and UBF1. (B) Lysates from various cell lines were immunoprecipitated (IP) with an anti-FLAG antibody, and the Western blots (WB) were successively developed with antibodies to either IRS-1 or FLAG. FLAG-tagged PES1 was detectable in both cells lines expressing it, but IRS-1 was visible only in 32D IGF-IR/IRS-1 cells. (C) Lysates from cells expressing either FLAG-tagged UBF1 (first two lanes) or FLAG-tagged PES1 (last two lanes) were immunoprecipitated with an anti-FLAG antibody, and the Western blot was developed with an antibody to UBF1.
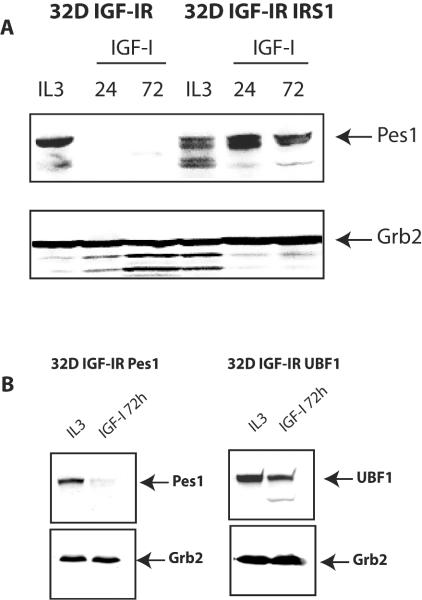
Levels of expression of PES1 in proliferating and differentiating 32D-derived cells.
The expression of endogenous UBF1 in 32D-derived cells has already been reported (61). Briefly, UBF1 is present abundantly in proliferating 32D IGF-IR and 32D IGF-IR/IRS-1 cells and disappears in differentiating cells. This finding is not surprising, as UBF1 is an exclusively nucleolar protein (67, 68) and the nucleolus involutes in terminally differentiated cells (50). We used an antibody to PES1 to detect endogenous PES1. PES1 expression in these cell lines was very much the same as UBF1 expression. In 32D IGF-IR cells, the expression of endogenous PES1 decreased rapidly, and by 48 h after a shift from IL-3 to IGF-I, it was no longer detectable (Fig. 8A). In contrast, in 32D IGF-IR/IRS-1 cells, the expression of PES1 continued at high levels at least for 96 h after the shift from IL-3 to IGF-I (Fig. 8A).
FIG. 8.
Expression of endogenous PES1 in 32D-derived cells. (A) 32D IGF-IR and 32D IGF-IR/IRS-1 cells were shifted from IL-3 to IGF-I, and the levels of endogenous PES1 expression in whole-cell lysates were measured. The time after the shift is indicated above the lanes in hours. PES1 expression persisted in transformed 32D IGF-IR/IRS-1 cells but not in differentiating 32D IGF-IR cells. (B) Expression of exogenous UBF1 and PES1 in differentiated 32D-derived cells. The cell lines examined were 32D IGF-IR cells overexpressing either PES1 or UBF1 3 days after a shift to IGF-1. Lanes IL-3 contained the same cells at zero time. Western blotting was done with antibodies to PES1 or UBF1. An anti-Grb2 antibody was used to monitor protein amounts.
Having established that endogenous PES1 and UBF1 are no longer expressed in differentiated 32D IGF-IR cells, we examined whether the exogenous proteins were still expressed in these cells. The results are shown in Fig. 8B. PES1 (endogenous or exogenous) disappears in cells induced to differentiate. However, contrary to what happens to endogenous UBF1 (61), exogenous UBF1 persists at least until day 4, although its expression is modestly decreased with respect to that at time zero. The persistence of exogenous UBF1 in 32D IGF-IR cells may explain why these cells are even larger than PES1 cells at day 6 after a shift to IGF-I (Fig. 5).
Effect of PES1 and UBF1 on the growth of MEFs.
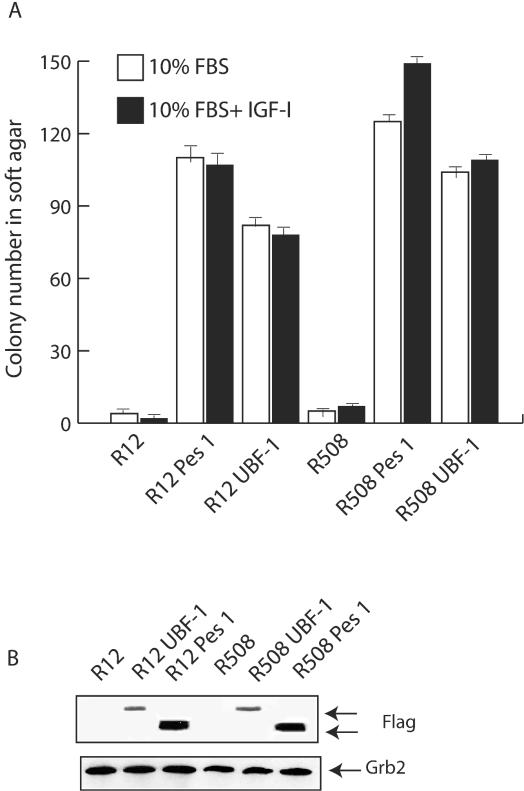
The results obtained thus far seem to suggest that both UBF1 and PES1 increase cell size and may affect cell proliferation but fail to make 32D-derived cells IL-3 independent. The results also suggest that PES1 and UBF1 cannot inhibit in 32D-derived cells the differentiation program initiated by IGF-IR (in cells that do not express IRS-1). If this suggestion is correct, then both UBF1 and PES1 should be able to transform MEFs that express IRS-1 and are known not to differentiate. For this purpose, we tested the effect of the ectopic expression of PES1 and UBF1 in MEFs that have substantial levels of IRS-1. We selected for this purpose R12 and R508 cells; these cells are derived from R− cells, which are MEFs originating from mouse embryos with a targeted disruption of the IGF-IR genes (34). Human IGF-IR was introduced into R12 and R508 cells to produce cell lines that express, respectively, 3 × 103 and 15 × 103 IGF-IRs/cell (52). R12 and R508 cells do not respond to IGF-I with proliferation, although they do respond to IGF-I with autophosphorylation of the receptor and modest tyrosyl phosphorylation of IRS-1 (49). Neither of these cell lines can form colonies in soft agar. We engineered both of these cell lines to express UBF1 or PES1 (see Methods and Materials), and we tested them for their ability to form colonies in soft agar. The results are shown in Fig. 9A, from which it is evident that both PES1 and UBF1 increased colony formation in soft agar of both R12 and R508 cells. As in previous experiments, parental R12 and R508 cells generated very few colonies in soft agar (averages of fewer than 1 in R12 cells and 2.5 in R508 cells). PES1 increased the number of colonies to 120 or more, while UBF1 increased the number of colonies to 70 to 110 per plate. Interestingly, the addition of IGF-I had no effect on R12 cells, while a small effect was noted for R508 cells. This finding is in agreement with previous reports indicating that R508 cells are slightly more sensitive to IGF-I than R12 cells (49, 52). Figure 9B shows the levels of expression of exogenous PES1 and UBF1 in MEFs.
FIG. 9.
PES1 and UBF1 induce the transformation of normal MEFs. The cells examined were parental R12 and R508 cells, which are 3T3-like MEFs incapable of forming colonies in soft agar (see the text). Both cell lines were infected with retroviruses expressing either PES1 or UBF1, and mixed populations were collected. (A) Parental and retrovirus-infected cells were tested for their ability to form colonies in soft agar. The ordinate shows the number of colonies after 3 weeks. The colonies counted measured at least 125 μm. Error bars indicate standard deviations. (B) Western blotting of lysates from infected cells expressing FLAG-tagged proteins.
DISCUSSION
We have investigated the effect of the overexpression of PES1 and UBF1 in a model of IGF-I-mediated cell differentiation and transformation. We selected the model of 32D murine myeloid cells for three reasons: (i) in these cells, IGF-IR sends a dual signal, one for differentiation and one for cell proliferation (64), as cells stimulated with IGF-I proliferate vigorously for 48 h before differentiating into granulocytes; (ii) ectopic expression of IRS-1 in these cells inhibits IGF-IR-induced differentiation and transforms the cells into tumor-forming cells (63); and (iii) IRS-1, like PES1 and UBF1, increases cell size (4, 45, 63), apparently through the activation of UBF1 (58, 62). With the premise that our conclusions must be interpreted within the context of the 32D-derived cell model, in which cells are modulated by IGF-IR signaling, we can point out some interesting observations.
First, we show that in 32D-derived cells, both UBF1 and PES1 can increase cell size, at least as well as IRS-1. Their effect on 32D cell size is evident in diverse growth conditions and is maintained even in differentiated cells. Second, PES1 alters the cell cycle distribution in exponentially growing 32D IGF-IR and 32D IGF-IR/IRS-1 cells and also slightly increases cell proliferation in the latter cell line. Third, despite their effect on cell size (and, for PES, on the cell cycle), neither of them can sustain the proliferation of 32D IGF-IR cells in the presence of IGF-I but in the absence of IL-3. 32D IGF-IR cells expressing PES1 or UBF1 stop growing, like their parental cells, after about 48 h. Their failure to grow further is accompanied by a failure to inhibit the IGF-I-mediated induction of differentiation, induction that is instead blocked by IRS-1 (63; this study). Fourth, however, PES1 and UBF1 transform (colony formation in soft agar) MEFs that do not have a differentiation program. These points are considered separately.
We selected PES1 and UBF1 because both of them play a role in ribosome biogenesis (12, 16, 31), which determines cell size (23, 37). IRS-1 also increases cell size (4, 69), and it may do so by binding and presumably activating UBF1 (58, 62). In addition, PES1 is also involved in DNA replication (12), thus connecting the increase in cell size with the cell cycle program, a requirement for normal cell division (13, 19). Our results confirm that PES1 and UBF1 can increase cell size. This finding was demonstrated by FACS analysis and by morphological observations. Neither PES1 nor UBF1 requires IRS-1 to increase cell size, as they increase cell size in 32D IGF-IR cells and parental 32D cells, which do not express IRS-1 (64, 70). This finding is compatible with the data in the literature (see above) indicating that IGF-IR and IRS-1 regulate, in a nonredundant way, only about 50% of cell size. The other 50% is IRS-1 independent.
PES1 is also known to affect DNA replication (12, 31), and our results are compatible with its role in this process. Exponentially growing cells had an altered cell cycle distribution, with a shortening of S phase and a lengthening of G2 phase, which resulted in overall growth in the first 48 h that was only slightly increased over that of parental cells. Interestingly, the effect of PES1 in these cells apparently is dependent on the activation of IGF-IR signaling. Cells growing in IL-3 do not seem to show the same dramatic changes in cell cycle distribution that are caused by the overexpression of PES1. A reasonable question at this point is whether cells in the G2 peak are truly in G2. Since at 24 h all cell lines are growing exponentially, with 98 to 99% of the cells in the cell cycle (44), FACS analysis ought to provide a true cell cycle distribution. This distribution persists at 6 days after a shift to IGF-I, by which time 32D IGF-IR cells are differentiated (the only exception being IRS-1-expressing cells). At 6 days, therefore, it would be meaningless to discuss a G2 phase; at that time, FACS analysis simply measures DNA amounts. It is intriguing that 32D IGF-IR cells expressing PES1 and UBF1 remain very large even when differentiated. Brown et al. (5) reported that HL60 cells halted in G1 or S phase differentiate normally. Morphological analysis of size in differentiated cells (6 days after a shift to IGF-I) was confirmed by FACS analysis (data not shown).
The most important conclusion of our experiments is that, of the three proteins that increase cell size (and, for PES1, also affect DNA replication), only IRS-1 can induce sustained cell proliferation in 32D IGF-IR cells. As mentioned above, 32D IGF-IR/IRS-1 cells not only are IL-3 independent but also actually form tumors in mice, something that parental 32D cells and 32D IGF-IR cells cannot do (63). The failure of UBF1 and PES1 to sustain IGF-1-dependent cell proliferation is accompanied by the differentiation of cells into granulocytes, as in parental 32D IGF-IR cells.
The differentiation of 32D IGF-IR cells, parental or overexpressing PES1 or UBF1, is documented unequivocally by morphological analysis, as they become granulocytes. It is confirmed by their inability to extinguish markers of differentiation that occur within the first 24 h after a shift to IGF-I, when 32D IGF-IR cells are still growing (44). These markers are MPO (47, 64) and 24p3 (46, 55) mRNAs and ID proteins. The ID proteins, especially ID1 and ID2, are strong inhibitors of differentiation (3, 27, 51). In 32D IGF-IR cells, ID2 gene expression is very low, almost undetectable. Ectopic expression of IRS-1 in these cells causes a dramatic increase in ID2 gene expression (2, 40, 47; this study). Neither UBF1 nor PES1 is capable of inducing ID2 expression in 32D IGF-IR cells.
A model to explain these results could be based on the hypothesis that PES1, UBF1, and IRS-1 all act on the cell cycle program and/or ribosome biogenesis but that only IRS-1 can extinguish the differentiation program initiated by activated IGF-IR. In this model, IRS-1 would send three different signals: (i) an increase in cell size (63); (ii) activation of the cell cycle program (49); and (iii) extinction of the differentiation program initiated by IGF-IR (32, 61, 63). This interpretation is supported by our finding that the overexpression of PES1 or UBF1 causes the transformation of MEFs. There is indeed no question that both PES1 and UBF1 can transform 3T3-like MEFs, as the number of colonies in soft agar is quite high. MEFs have no propensity to differentiate, and PES1 and UBF1 can be transforming for these cells because they do not need to extinguish a differentiation program. It seems a sensible explanation that for 32D-derived cells that also have a differentiation program, this latter has to be blocked for sustained growth to occur.
We mentioned above that IRS-1 induces ID2 expression, which could explain simply why it blocks differentiation. ID proteins bind to pRb (20, 29), which binds to C/EBPα (8) and induces differentiation. It is possible, though, that IRS-1 has alternative pathways for inhibit diffentiation. pRb also inhibits RNA polymerase I transcription by binding and inactivating UBF1 (7, 9, 17, 18, 69), while IRS-1 binds UBF1 and activates transcription from the ribosomal DNA promoter (46, 58). Alternatively, IRS-1 could displace inhibitory pRb from UBF1. It is also possible that different pathways merge. Rockman et al. (51) reported that β-catenin up-regulates ID2 expression, and we have found that nuclear IRS-1 interacts with β-catenin (unpublished data).
In conclusion, our results indicate that PES1 and UBF1 cannot induce sustained cell proliferation in 32D IGF-IR cells, despite their effects on cell size and DNA replication, which allow them to transform MEFs in cultures. We propose that PES1 and UBF1 are incapable of extinguishing the differentiation program that is initiated in 32D IGF-IR cells simultaneously with the induction of the cell cycle program. Although there are other explanations, it is important to point out that IGF-IR in these cells already sends a mitogenic signal, as the cells grow vigorously for 48 h, so that the inhibition of differentiation really may be the crucial event for sustained cell proliferation. In the absence of a differentiation signal, as in MEFs, PES1 and UBF1 can transform contact-inhibited cells into cells capable of forming colonies in soft agar. This interpretation is supported by the report that c-Myc, which is required for the initiation of the cell cycle program (22), is induced by IGF-I (49). Finally, these results support the concept proposed by several investigators that a block in differentiation is an integral part of malignancies, particularly hemopoietic malignancies (14, 42, 60). Indeed, our data provide a good model for studying the molecular mechanisms that determine the outcome of differentiation and transformation in myeloid cells.
Acknowledgments
This work was supported by grants CA78890 and CA56309 from the National Institutes of Health.
REFERENCES
- 1.Arkins, S., N. Rebeiz, D. L. Brunke-Reese, C. Minshall, and K. W. Kelley. 1995. The colony-stimulating factors induce expression of insulin-like growth factor-I messenger ribonuclei acid during hematopoiesis. Endocrinology 136:1153-1160. [DOI] [PubMed]
- 2.Belletti, B., M. Prisco, A. Morrione, B. Valentinis, M. Navarro, and R. Baserga. 2001. Regulation of Id2 gene expression by the IGF-I receptor requires signaling by phosphatidylinositol-3 kinase. J. Biol. Chem. 276:13867-13874. [DOI] [PubMed] [Google Scholar]
- 3.Benezra, R., R. L. Davis, D. Lockshon, D. L. Turner, and H. Weintraub. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49-59. [DOI] [PubMed] [Google Scholar]
- 4.Bohni, R., J. Riesco-Escovar, S. Oldham, W. Brogiolo, H. Stocker, B. F. Andruss, K. Beckingham, and E. Hafen. 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS-1. Cell 97:865-875. [DOI] [PubMed] [Google Scholar]
- 5.Brown, G., M. T. Drayson, J. Durham, K. M. Toellner, P. J. Hughes, M. A. Choudhry, D. R. Taylor, and R. H. Michell. 2002. HL60 cells halted in G1 or S phase differentiate normally. Exp. Cell Res. 281:28-38. [DOI] [PubMed] [Google Scholar]
- 6.Castaigne, S., C. Chomienne, M. T. Daniel, P. Ballerini, R. Berger, P. Fenaux, and L. Degos. 1990. All trans retinoic acid as a differentiation therapy for acute promyelocytic leukemia. I. Clinical results. Blood 76:1704-1709. [PubMed] [Google Scholar]
- 7.Cavanaugh, A. H., W. M. Hempel, L. J. Taylor, V. Rogalsky, G. Todorov, and L. I. Rothblum. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374:177-180. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P. L., D. J. Riley, Y. Chen, and L. W. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 9.Ciarmatori, S., P. H. Scott, J. E. Sutcliffe, A. McLees, H. M. Alzuherri, J. H. Dannenberg, H. Riele, L. Grummt, R. Voit, and R. J. White. 2001. Overlapping function of the pRb family in the regulation of rRNA synthesis. Mol. Cell. Biol. 21:5806-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comai, L., Y. Song, C. Tan, and T. Bui. 2000. Inhibition of RNA polymerase I transcription in differentiated myeloid leukemia cells by inactivation of selectivity factor-1. Cell Growth Differ. 11:63-70. [PubMed] [Google Scholar]
- 11.Datta, P. K., S. Budhiraja, R. R. Reichel, and T. J. Samson. 1997. Regulation of ribosomal RNA gene transcription during retinoic acid-induced differentiation of mouse teratocarcinoma cells. Exp. Cell Res. 231:198-205. [DOI] [PubMed] [Google Scholar]
- 12.Du, Y. C. N., and B. Stillman. 2002. Yph1p, an ORC-interacting protein: potential link between cell proliferation control, DNA replication and ribosome biogenesis. Cell 109:835-848. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, R. S. S., and P. Nurse. 1978. Novel cell cycle control of RNA synthesis in yeasts. Nature 27:726-730. [DOI] [PubMed] [Google Scholar]
- 14.Gilliland, D. G., and M. S. Tallman. 2002. Focus on acute leukemias. Cancer Cell 1:417-420. [DOI] [PubMed] [Google Scholar]
- 15.Grueneberg, D. A., L. Pablo, K. Q. Hu, P. August, Z. Weng, and J. Papkoff. 2003. A functional screen in human cells identifies UBF2 as an RNA polymerase II transcription factor that enhances the β-catenin signaling pathway. Mol. Cell. Biol. 23:3936-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grummt, I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62:109-153. [DOI] [PubMed] [Google Scholar]
- 17.Hannan, K. M., R. D. Hannan, S. D. Smith, L. S. Jefferson, M. Lun, and L. I. Rothblum. 2000. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 19:4988-4999. [DOI] [PubMed] [Google Scholar]
- 18.Hannan, K. M., B. K. Kennedy, A. H. Cavanaugh, R. D. Hannan, I. Hirschler-Laszkiewicz, L. S. Jefferson, and L. I. Rothblum. 2000. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene 19:3487-3497. [DOI] [PubMed] [Google Scholar]
- 19.Hartwell, L. H. 1978. Cell division from a genetic perspective. J. Cell Biol. 77:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iavarone, A., P. Garg, A. Lasorella, J. Hsu, and M. A. Israel. 1994. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 8:1270-1284. [DOI] [PubMed] [Google Scholar]
- 21.Iben, S., H. Tschochner, M. Bler, D. Hoogstraten, P. Hozak, J. M. Egly, and I. Grummt. 2002. TFIIH plays an essential role in RNA polymerase I transcription. Cell 109:297-306. [DOI] [PubMed] [Google Scholar]
- 22.Johansen, L. M., A. Iwama, T. A. Lodie, K. Sasaki, T. W. Felsher, T. R. Golub, and D. G. Tenen. 2001. c-Myc is a critical target for C/EBPα in granulopoiesis. Mol. Cell. Biol. 21:3789-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz, and M. Tyers. 2002. Syst. identification of pathways that couple cell growth and division in yeast. Science 297:395-400. [DOI] [PubMed] [Google Scholar]
- 24.Keeshan. K., G. Santilli, F. Corradini, D. Perrotti, and B. Calabretta. 2003. Transcription activation function of C/EBPα is required for induction of granulocytic differentiation. Blood 102:1267-1275. [DOI] [PubMed] [Google Scholar]
- 25.Kim, B., P. S. Leventhal, M. F. White, and E. L. Feldman. 1998. Differential regulation of insulin receptor substrate-2 and mitogen-activated protein kinase tyrosine phosphorylation by phosphatidylinositol 3-kinase inhibitors in SH-SY5Y human neuroblastoma cells. Endocrinology 139:4881-4889. [DOI] [PubMed] [Google Scholar]
- 26.Kinoshita, Y., A. D. Jarell, J. M. Flaman, G. Foltz, J. Schuster, B. L. Sopher, D. K. Irvi, H. I. Kornblum, P. S. Nelson, P. Hoieter, and R. S. Morrison. 2001. Pescadillo, a novel cell cycle regulatory protein abnormally expressed in malignant cells. J. Biol. Chem. 276:6656-6665. [DOI] [PubMed] [Google Scholar]
- 27.Kreider, B. L., R. Benezra, G. Rovera, and T. Kadesch. 1992. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science 255:1700-1702. [DOI] [PubMed] [Google Scholar]
- 28.Larson, D. E., W. Q. Xie, M. Glibetic, D. O'Mahony, B. H. Sells, and L. I. Rothblum. 1993. Coordinated decrease in rRNA gene transcription factors and rRNA synthesis during muscle differentiation. Proc. Natl. Acad. Sci. USA 90:7933-7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasorella, A., M. Noseda, M. Beyna, and A. Iavarone. 2000. Id2 is a retinoblasoma protein target and mediates signaling by Myc oncoproteins. Nature 407:592-598. [DOI] [PubMed] [Google Scholar]
- 30.Leevers, S. J., D. Weinkove, L. K. MacDougall, E. Hafen, and M. D. Waterfield. 1996. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15:6584-6594. [PMC free article] [PubMed] [Google Scholar]
- 31.Lerch-Gaggl, A., J. Haque, J. Li, G. Ning, P. Traktman, and S. A. Duncan. 2002. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 277:45347-45355. [DOI] [PubMed] [Google Scholar]
- 32.Li, Y. M., D. H. Schacher, Q. Liu, S. Arkins, N. Rebeiz, R. H. McCusker, Jr., R. Dantzer, and K. W. Kelley. 1997. Regulation of myeloid growth and differentiation by the insulin-like growth factor receptor. Endocrinology 138:362-368. [DOI] [PubMed] [Google Scholar]
- 33.Lin, C. Y., J. A. Tuan, P. Scalia, T. Bul, and L. Comai. 2002. The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr. Biol. 12:2142-2146. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J.-P., J. Baker, A. S. Perkins, E. J. Robertson, and A. Efstratiadis. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (igf-1) and type 1 IGF receptor (Igf1r). Cell 75:59-72. [PubMed] [Google Scholar]
- 35.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila 6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 36.Morrione, M., M. Navarro, G. Romano, M. Dews, K. Reiss, B. Valentinis, B. Belletti, and R. Baserga. 2001. The role of the insulin receptor substrate-1 in the differentiation of rat hippocampal neuronal cells. Oncogene 20:4842-4852. [DOI] [PubMed] [Google Scholar]
- 37.Moss, T., and V. Y. Stefanovsky. 1995. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acids Res. Mol. Biol. 50:25-66. [DOI] [PubMed] [Google Scholar]
- 38.Mueller, C., M. Alunni-Fabbroni, E. Kowenz-Leutz, X. Mo, M. Tommasino, and A. Leutz. 1999. Separation of C/EBPα-mediated proliferation arrest and differentiation pathways. Proc. Natl. Acad. Sci. USA 96:7276-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro, M., and R. Baserga. 2001. Limited redundancy of survival signals from the type 1 insulin-like growth factor receptor. Endocrinology 142:1073-1081. [DOI] [PubMed] [Google Scholar]
- 40.Navarro, M., B. Valentinis, B. Belletti, G. Romano, K. Reiss, and R. Baserga. 2001. Regulation of Id2 gene expression by the type 1 IGF receptor and the insulin receptor substrate-1. Endocrinology 142:5149-5157. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien, T., and R. Tjian. 2000. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc. Natl. Acad. Sci. USA 97:2456-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabst, T., B. U. Mueller, P. Zhang, H. S. Radomska, S. Narravula, S. Schnittger, G. Behre, W. Hiddemann, and D. G. Tenen. 2001. Dominant negative mutations of CEBPα, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 27:263-270. [DOI] [PubMed] [Google Scholar]
- 43.Perrotti, D., V. Cesi, R. Trotta, C. Guerzoni, G. Santilli, K. Campbell, A. Iervolino, F. Condorelli, C. Gambacorti-Passerini, M. A. Caligiuri, and B. Calabretta. 2001. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat. Genet. 30:48-58. [DOI] [PubMed] [Google Scholar]
- 44.Peruzzi, F., M. Prisco, M. Dews, P. Salomoni, E. Grassilli, G. Romano, B. Calabretta, and R. Baserga. 1999. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol. Cell. Biol. 19:7203-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pete, G., G. R. Fuller, J. M. Oldham, D. R. Smith, A. J. D'Ercole, C. R. Kahn, and P. K. Lund. 1999. postnatal growth responses to insulin-like growth factor 1 in insulin receptor substrate-1 mice. Endocrinology 140:5478-5487. [DOI] [PubMed] [Google Scholar]
- 46.Prisco, M., F. Santini, R. Baffa, M. Liu, R. Drakas, A. Wu, and R. Baserga. 2002. Nuclear translocation of IRS-1 by the SV40 T antigen and the activated IGF-I receptor. J. Biol. Chem. 277:32078-32085. [DOI] [PubMed] [Google Scholar]
- 47.Prisco, M., F. Peruzzi, B. Belletti, and R. Baserga. 2001. Regulation of Id gene expression by the type 1 insulin-like growth factor: roles of Stat3 and the tyrosine 950 residue of the receptor. Mol. Cell. Biol. 21:5447-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeder, R. H. 1999. Regulation of RNA polymerase transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 62:293-327. [DOI] [PubMed] [Google Scholar]
- 49.Reiss, K., B. Valentinis, X. Tu, S. Q. Xu, and R. Baserga. 1998. Molecular markers of IGF-I-mediated mitogenesis. Exp. Cell Res. 242:361-372. [DOI] [PubMed] [Google Scholar]
- 50.Ringertz, N. R., and R. E. Savage. 1976. Cell hybrids. Academic Press, Inc., New York, N.Y.
- 51.Rockman, S. P., S. A. Currie, M. Ciavarella, E. Vincan, C. Dow, R. J. S. Thomas, and W. A. Phillips. 2001. Id2 is a target of the β-catenin/T cell factor pathway in colon carcinoma. J. Biol. Chem. 276:45113-45119. [DOI] [PubMed] [Google Scholar]
- 52.Rubini, M., A. Hongo, C. D'Ambrosio, and R. Baserga. 1997. The IGF-I receptor in mitogenesis and transformation of mouse embryo cells: role of receptor number. Exp. Cell Res. 230:284-292. [DOI] [PubMed] [Google Scholar]
- 53.Sadowski, C. L., T. S. Choi, M. Le, T. T. Wheeler, L. H. Wang, and H. B. Sadowski. 2001. Insulin induction of SOCS-2 and SOCS-3 mRNA expression in C2C12 skeletal muscle cells is mediated by Stat5. J. Biol. Chem. 276:20703-20710. [DOI] [PubMed] [Google Scholar]
- 54.Sawyers, C. L., C. T. Denny, and O. N. Witte. 1991. Leukemia and the disruption of normal hematopoiesis. Cell 64:337-350. [DOI] [PubMed] [Google Scholar]
- 55.Sciacca, L., M. Prisco, A. Wu, A. Belfiore, R. Vigneri, and R. Baserga. 2003. Signaling differences from the Α and B isoforms of the insulin receptor (IR) in 32D cells in the preence or absence of IR substrate-1. Endocrinology 144:2650-2658. [DOI] [PubMed] [Google Scholar]
- 56.Sell, C., M. Rubini, R. Rubin, J.-P. Liu, A. Efstratiadis, and R. Baserga. 1993. Simian virus 40 large tumor antigen is unable to transform mouse embryonic fibroblasts lacking type-1 IGF receptor. Proc. Natl. Acad. Sci. USA 90:11217-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70S6K/p85S6K gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, H., X. Tu, M. Prisco, A. Wu, I. Casiburi, and R. Baserga. 2003. Insulin-like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol. Endocrinol. 17:472-486. [DOI] [PubMed] [Google Scholar]
- 59.Sun, X. J., S. Pons, L. M. Wang, Y. Zhang, L. Yenush, D. Burks, M. G. Myers, Jr., E. Glasheen, N. G. Copeland, N. A. Jenkins, and M. F. White. 1997. The IRS-2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Mol. Endocrinol. 11:251-262. [DOI] [PubMed] [Google Scholar]
- 60.Tenen, D. G. 2001. Abnormalities of CEBP alpha transcription factor: a major target in acute myeloid leukemia. Leukemia 15:688-689. [DOI] [PubMed] [Google Scholar]
- 61.Tu, X., R. Baffa, S. Luke, M. Prisco, and R. Baserga. 2003. Intracellular redistribution of nuclear and nucleolar proteins during differentiation of 32D murine hemopoietic cells. Exp. Cell Res. 288:119-130. [DOI] [PubMed] [Google Scholar]
- 62.Tu, X., P. Batta, N. Innocent, M. Prisco, I. Casaburi, B. Belletti, and R. Baserga. 2002. Nuclear translocation of insulin receptor substrate-1 by oncogenes and IGF-I: effect on ribosomal RNA synthesis. J. Biol. Chem. 277:44357-44365. [DOI] [PubMed] [Google Scholar]
- 63.Valentinis, B., M. Navarro, T. Zanocco-Marani, P. Edmonds, J. McCormick, A. Morrione, A. Sacchi, G. Romano, K. Reiss, and R. Baserga. 2000. Insulin receptor substrate-1, p70S6K and cell size in transformation and differentiation of hemopoietic cells. J. Biol. Chem. 275:25451-25459. [DOI] [PubMed] [Google Scholar]
- 64.Valentinis, B., G. Romano, F. Peruzzi, A. Morrione, M. Prisco, S. Soddu, B. Cristofanelli, A. Sacchi, and R. Baserga. 1999. Growth and differentiation signals by the insulin-like growth factor 1 receptor in hemopoietic cells are mediated through different pathways. J. Biol. Chem. 274:12423-12430. [DOI] [PubMed] [Google Scholar]
- 65.Valtieri, M., D. J. Tweardy, D. Caracciolo, K. Johnson, F. Mavilio, S. Altmann, D. Santoli, and G. Rovera. 1987. Cytokine dependent granulocytic differentiation. J. Immunol. 138:3829-3835. [PubMed] [Google Scholar]
- 66.Verdu, J., M. A. Buratovich, E. L. Wilder, and M. J. Birnbaum. 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1:500-506. [DOI] [PubMed] [Google Scholar]
- 67.Voit, R., M. Hoffmann, and I. Grummt. 1999. Phosphorylation of G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voit, R., A. Kuhn E. E. Sander, and I. Grummt. 1995. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23:2593-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voit, R., K. Schafer, and I. Grummt. 1997. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol. 17:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, L. M., M. G. Myers, Jr., X. J. Sun, S. A. Aaronson, M. White, and J. H. Pierce. 1993. IRS-1: essential for insulin- and IL-4-stimulated mitogenesis in hemopoietic cells. Science 261:1591-1594. [DOI] [PubMed] [Google Scholar]
- 71.Ward, A. C., L. Smith, J. P. de Koning, Y. van Aesch, and I. P. Touw. 1999. Multiple signals mediate proliferation, differentiation and survival from the granulocyte-colony stimulating factor receptor in myeloid 32D cells. J. Biol. Chem. 274:14956-14962. [DOI] [PubMed] [Google Scholar]
- 72.White, M. F. 1998. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell. Biochem. 182:3-11. [PubMed] [Google Scholar]
- 73.Zamorano, J., H. Y. Wang, L.-M., Wang, J. H. Pierce, and A. D. Keegan. 1996. IL-4 protects cells from apoptosis via the insulin receptor substrate pathway and a second independent signaling pathway. J. Immunol. 157:4926-4934. [PubMed] [Google Scholar]
- 74.Zhou-Li, F., S.-Q. Xu, M. Dews, and R. Baserga. 1997. Cooperation of simian virus 40 T antigen and insulin receptor substrate-1 in protection from apoptosis induced by interleukin-3 withdrawal. Oncogene 15:961-970. [DOI] [PubMed] [Google Scholar]