Abstract
J-chain (Joining chain) is a small polypeptide that regulates multimerization of secretory IgM and IgA, the only two mammalian Igs capable of forming multimers. J-chain also is required for polyIg-receptor (pIgR)-mediated transport of these Ig classes across the mucosal epithelium. It is generally assumed that all plasma cells express J-chain regardless of expressed isotype, despite the documented presence of J-chain-negative plasma cells in mammals, specifically in all monomeric IgA- and some IgG-secreting cells. Compared to most other immune molecules, J-chain has not been extensively studied, due, in part, to technical limitations. Even the reported phenotype of the J-chain knockout mouse is often misunderstood or underappreciated. In this short review, we will discuss J-chain in light of the various proposed models of its expression and regulation, with an added focus on the evolutionary significance of J-chain and its expression in different B cell lineages/differentiation states.
Joining Chain (J-chain)
Joining chain, or J-chain, is a small polypeptide that regulates the multimerization of IgM and IgA. It appeared with the emergence of adaptive immunity in jawed vertebrates (1), and is an unusual molecule, in that it does not appear to be a member of any characterized protein domain family (2). Mammalian J-chain is acidic and contains eight cysteine residues, six of which form intrachain disulfide bonds (C1–C6, C4–C5, and C7–C8), while the remaining two form interchain disulfide bonds with cysteines in the IgM or IgA heavy-chain tails (2, 3). When associated with J-chain, mammalian IgM is secreted as a pentamer and IgA as a dimer, the typical form in mucosal secretions (Table I). In the absence of J-chain IgA is secreted as a monomer (Table I), the form most common in the blood (4). This monomeric, J-chain− IgA is secreted from different cells from those producing IgA dimers, and the two forms of IgA have distinctive functions, such as providing a barrier to commensal infections (dimeric IgA) and induction of inflammation (monomeric IgA) (5, 6). The other mammalian isotypes IgG/E/D do not multimerize (Table I), although some plasma cells that express these isotypes also express J-chain. Due to differences in the secretory tail of the IgG/E/D heavy chains, J-chain does not associate with these isotypes (discussed below in the context of all vertebrate Igs), hence their secretion as monomers regardless of J-chain expression (7).
Table I.
Summary of Ig isotypes with a focus on multimerized and mucosally secreted isotypes throughout evolution
| J-chain present in genome |
Monomeric isotypes |
Multimerized Ig isotypes |
Normal multimerization states |
J-chain associated |
Muscosal transport via pIgR |
|
|---|---|---|---|---|---|---|
| Mammals | Yes | IgD, IgG, IgE | IgM | Pentamers | Yes | Yes |
| IgA1 | Monomers | No | No | |||
| Dimers | Yes | Yes | ||||
| Bony fish | No2 | IgD | IgM | Tetramers | No | Yes3 |
| IgT | Monomers | No | No | |||
| Tetramers (non-covalent) | No | Yes3 | ||||
| Amphibians | Yes | IgD, IgY | IgM | Hexamers, pentamers | Yes | Yes |
| IgX | Hexamers, perntamers | No | ?4 | |||
| Cartilaginous fish | Yes | IgW?, IgNAR | IgM1, IgW? | Monomers | No | Unlikely |
| Pentamers | Yes | Unlikely |
Some isotypes such as IgA in mammals and IgM in cartilaginous fish can be secreted in either multimeric or monomeric forms, with or without J-chain, respectively.
While J-chain has been lost in teleost fish, it has been found in a lobed-fin fish, the lungfish, which is closely related to tetrapods.
Although teleosts do not express J-chain, IgT has been shown to interact with the pIgR secretory component directly.
The mechanism of mucosal transport without J-chain association in these cases is unclear.
In addition to multimerization, J-chain is required for Ig transport across the mucosal epithelium in tetrapods (8). The C-terminal domain of J-chain is required for association with a portion of the poly-Ig receptor (pIgR) known as secretory component (SC) or ‘secretory piece.’ While J-chain is associated with Ig in plasma cells, epithelial cells produce pIgR. Secreted J-chain+ Igs bind to pIgR on the basolateral surface of mucosal epithelial cells, leading to endocytosis of the entire complex and transport by transcytosis to the cell’s apical surface. Proteolytic cleavage then releases Ig into the lumen (9). The SC remains with J-chain and IgM/A after transcytosis across the epithelium (10).
Based on current understanding of J-chain transcriptional control, discussed in detail below, it is widely assumed that all plasma cells express J-chain (11–13), although there are inconsistencies in the documentation of J-chain protein levels in mammalian plasma cells, including the monomeric IgA-secreting cells (14). The connection between RNA expression and protein levels is not always clear, although they are often used interchangeably in the literature. Some of this confusion stems from technical difficulties in studying the molecule. Structural changes, or epitope masking of J-chain when it is associated with Ig, have made the detection of J-chain, and the production of high-quality antibodies that recognize native J-chain, challenging. These difficulties are exemplified by inconsistencies in J-chain detection in early studies. Whether or not a denaturant (e.g. urea) was used to “unmask” J-chain epitopes before immunohistochemical detection led to different interpretations of J-chain expression in plasma cells. Specifically, studies that did not utilize urea treatment before staining for J-chain inadvertently underestimated the number of J-chain+ cells (5, 15, 16).
The unique structure and biochemical behavior of J-chain, combined with its lack of association with monomeric Igs such as IgG, have relegated the J-chain to a lower echelon of immune molecules and it has been relatively overlooked for many years, since its heyday in the 1970s and ‘80s, especially in the lab of the esteemed Marian Koshland (17), and a short resurgence over a decade ago with the report of the J chain knockout mouse by Leanderson’s group. Several basic questions about J-chain gene regulation and expression remain unanswered, and there are key differences in the two central models of J-chain expression. In this review, we hope to highlight and re-define some of these issues and put the J-chain back onto the radar.
J-chain knockout (KO) confusion
While mouse IgM is secreted as a pentamer in the presence of J-chain, in its absence, IgM forms disordered oligomers rather than monomers, like IgA (7, 18–21). This KO phenotype is frequently misquoted as producing hexameric IgM from J-chain-null cells, when in fact IgM was described as being secreted as an “oligomeric form of undefined structure” in an early KO mouse study (19). Cell culture studies of IgM/J-chain expression, however, have provided different results from the KO mouse (18–20), which may explain some of the confusion about IgM hexamers. The absence of J-chain does lead to secretion of high levels of hexameric IgM in in vitro cell-based models (22), however even in some of these cellular studies both tetrameric and higher molecular weight species were found in addition to hexameric and pentameric IgM (23). Additionally, although hexameric IgM secreted from J-chain-negative cells is superior to the pentamer in complement activation (24), IgM from J-chain KO mice was impaired in complement activation (19), supporting the conclusion that J-chain KO mice actually produce very little hexameric IgM. Presumably there are other factors at play in the secretion of IgM in J-chain− plasma cells in vivo compared to cell culture systems, and accordingly we believe the KO mice likely provide a better representation of a J-chain-null IgM environment in mammals.
Although J-chain-negative, hexameric IgM was not the predominant species in the J-chain KO mouse, somewhat surprisingly, hexameric IgM lacking J-chain has been described in normal human sera (25), and is associated with human antibody-related diseases such as Waldenström’s macroglobulinemia, a B cell lymphoma, and cold agglutinin disease (26–28). Additionally, in women vaccinated to uropathogenic bacteria, those that responded to the vaccination had normal levels of pentameric IgM, whereas non-responders had increases in hexameric IgM (29). It is important to note that, as in the KO mouse, IgM multimers consisting of tetramers and oligomers were also described, along with hexameric IgM, in the J-chain negative fraction from patients with Waldenström’s macroglobulinemia (27), again suggesting that the KO mouse is a physological model of J-chain’s characteristics in humans.
Unconventional J-chain expression in non-B cells
Other complications in examining J-chain regulation have arisen. As mentioned, J-chain traditionally has been associated only with Ig mulimerization and secretion; however, both B and T cells can express J-chain early in development (30), and J-chain expression has also been described in a subset of dendritic cells (DC) (31). These J-chain+ DCs are CD11c+ and produce indoleamine 2,3-dioxygenase (IDO), an important tolerogenic DC signal. Both IDO and CD11c+ DCs are decreased in J-chain KO mice (32), however it is unclear how J-chain expression is regulated in these cells or how/why IDO production is upregulated. Additionally, J-chain KO mice are deficient in B cell memory, and unexpectedly, also have compromised T helper cell function, although J-chain expression is extinguished in mature T cells (30). In fact, this defective B cell memory phenotype is suspected to be dependent on T cells rather than B cells, based on the finding that mice receiving primed T cells, but not B cells, from a J-chain−/− mouse had a diminished antigen-specific Ig response after transfer (33). Finally, J-chain mRNA was clearly detected in lungfish intestinal epithelial cells, with no speculation on its function (34). These preliminary studies of J-chain expression and function in non-B cells demonstrate that there is more to the biology of J-chain than just Ig secretion. The question remains whether J chain truly has a role these cells (30, 31, 33), or whether it is simply upregulated in some cells, without regard to function, as a consequence of the transcriptional program and/or chromatin state in these cells.
Evolutionary significance of J-chain and mucosal isotypes
IgM is present in almost all jawed vertebrates from shark to human (35, 36), while mucosal IgA is found in all tetrapods (named IgX in amphibians (37, 38)). Teleosts (bony fish) also have a dedicated mucosal Ig class named IgT, which is unrelated to IgA (39, 40) (Table I). J-chain arose in the earliest jawed vertebrates (e.g. placoderms and cartilaginous fish, see below; note, one study suggesting J chain’s presence in earthworms could not be verified in any other invertebrate or agnathan species (41)), but surprisingly was lost from teleosts, although it has been described in a lobed-fin fish, the lungfish (34). Secretory B cells and B cell-specific transcription factors have been well studied in teleosts, and despite their lack of J-chain, plasma cells nevertheless produce multimerized IgM and IgT that can be transported across epithelia via pIgR (39, 42–44) (Table I). Interestingly, Xenopus mucosal IgX also multimerizes in the absence of J-chain, although unlike teleosts, J-chain is present in frogs and associates with IgM heavy chains (37) (Table I). This dichotomy raises interesting questions about the regulation of J-chain in Xenopus. The secretory tail of IgM and IgA is remarkably well conserved in phylogeny in its length, glycosylation site, and presence of cysteine as the penultimate residue that bonds covalently to J-chain (45). The IgX secretory tail has all of these features except the conserved cysteine and therefore does not associate with J chain (37, 46). However, questions remain about whether J-chain is still expressed, but simply does not associate, in IgX-expressed plasma cells (as suggested for mammalian IgG), or if it is regulated transcriptionally (Table I). Since transcription factors thought to be important in J-chain regulation are well conserved throughout vertebrate evolution (47, 48), studies of J-chain expression in plasma cells of non-mammalian models might aid in the understanding of J-chain regulation in all vertebrates (see below). Additionally, regarding function, it would be important to know if IgX is secreted and maintained in the lamina propria (LP) for defense, perhaps like monomeric IgA, or if it can be transcytosed into the lumen without J-chain association, like bony fish IgT.
Even in mammals, the number of IgA genes varies in different species, complicating study of their regulation and association with J-chain. Humans have two IgA genes, IgA1 and IgA2, while other species such as rabbit have many more (49). In humans IgA2 preferentially associates with J-chain (50), and class switch from IgA1 to IgA2 has been documented to occur in peripheral tissues such as the colon, often in a T-independent fashion (51); however, the transcriptional regulation of these two forms is not well understood. In contrast, mice only possess one IgA gene and thus secretion of monomeric vs. multimeric IgA is regulated solely via J-chain association. Therefore, despite the discrepancy between the mouse KO and human serum expression regarding IgM multimerization, the mouse provides a useful and straightforward system to study J-chain expression in mucosal, IgA-secreting cells.
In addition, basic J-chain regulation for Ig monomer secretion must also occur in other species such as cartilaginous fish, which unlike teleosts, do have the J-chain, and use it (perhaps exclusively) for IgM multimerization. Sharks do not express IgA, or any known dedicated mucosal isotype, but like IgA-secretion in mammals, shark plasma cells secrete IgM in two forms, as monomers or as canonical pentamers (52–54) (Table I). Similar to the situation with mammalian IgA, J-chain is only present in the multimeric IgM form (Table I) (1, 55, 56), and therefore its regulation must be tightly controlled to permit secretion of monomeric IgM. As discussed for Xenopus above, further study of shark plasma cells may shed light on J-chain gene regulation in all vertebrates. Since J-chain expression in shark pentameric IgM secreting cells and lack thereof in monomeric secretors is so clear cut, sharks may provide a less ambiguous system for studying Ig multimerization and J-chain control.
Transcriptional regulation of J-chain
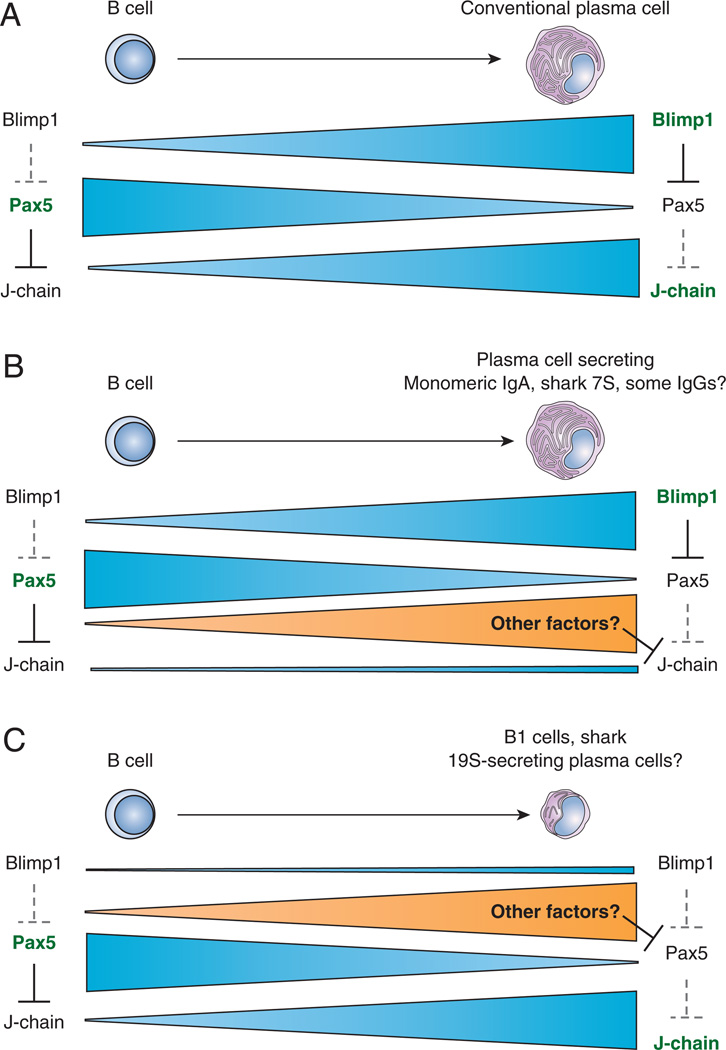
J-chain expression is activated by B-MEF2 and repressed by Pax5 (11, 12). Pax5 is considered the master regulator transcription factor of the B cell lineage (57, 58), because it suppresses expression of non-B cell genes (13, 59–61), including those required for plasma cell identity. Upon differentiation into Ig-secreting cells, Blimp1, the master regulator of plasma cell development, downregulates Pax5, resulting in de-repression of genes required for Ig secretion and the plasma cell phenotype, including J-chain (11, 61–63) (Figure 1A). Thus, the current appreciation of B cell transcriptional networks dictates that J-chain transcript should be expressed when Pax5 is decreased in all plasma cells, regardless of the isotype expressed (10) (Figure 1A).
Figure 1. Expression of main B cell and plasma cell transcription factors as they relate to J-chain expression based on current transcriptional networks.
Canonical Pax5 and Blimp1 expression shown in (A): normally, B cells (left) have high levels of Pax5, which binds to the J-chain promoter to prevent expression of J-chain. In contrast, Blimp1 is highly expressed in plasma cells (right), where it represses Pax5 expression, and therefore indirectly allows for J-chain expression. Grey dotted lines indicate relief of control due to absence of repressor. Bold factors in green text indicate expression at that stage. (B) Mammalian monomeric IgA-secretors and shark 7S IgM secretors, however, may use other unknown factors in the repression of J-chain, since, despite the absence of Pax5 in secreting cells, J-chain must not be expressed in order for these isotypes to be secreted as monomers. (C) Some cells, such as mammalian B1 cells and shark 19S IgM have been suggested to secrete Ig, and express J-chain, in the absence of Blimp1 (70, 100), so other factors might also control Pax5 expression in these cells as well. Of note, these Blimp1-negative secreting cells may secrete Ig at lower levels, indicated here by presence of fewer cisternae in (C).
The J-chain promoter is similar to the Igκ promoter (64) in that both contain an octamer element, a penta-deca element, and an E-box motif 5' of the TATA box (64). Three control elements have also been described, JA, JB, and JC. JA is bound by B-MEF2, PU.1 binds to JB, and Pax5 binds element JC in a manner that blocks binding to JA (11, 12, 21). Pax5 thus represses J-chain by forming a steric barrier to prevent binding of positive regulator B-MEF2 (12). Despite these similarities between the Igκ and J-chain promoters, J-chain is not expressed in the same cells or stages as Igκ, so other unidentified elements must be essential for J-chain expression. Filling this gap would provide crucial information on distinguishing between the models of plasma cell differentiation outlined below.
As mentioned, this canonical transcriptional model holds that J-chain should be expressed in all plasma cells when Pax5 repression is relieved (Figure 1A). Therefore, it has been presumed that it is expressed but quickly degraded in many cells secreting isotypes such as IgG, that do not associate with J-chain or use it for multimerization (65, 66) (Figure 2A). This degradation model is supported by studies showing that the amount of J-chain protein correlates with the amount of RNA present in Ig-secreting cells (67), and, in myeloma lines, unsecreted J-chain is degraded (65). While simple degradation makes sense for IgG+ cells, more experimental support for this paradigm is required, since this theory neither accounts for J-chain− monomeric IgA secretors (Figure 1B), nor clarifies why J-chain protein is found in some but not all IgG-secreting cells (68).
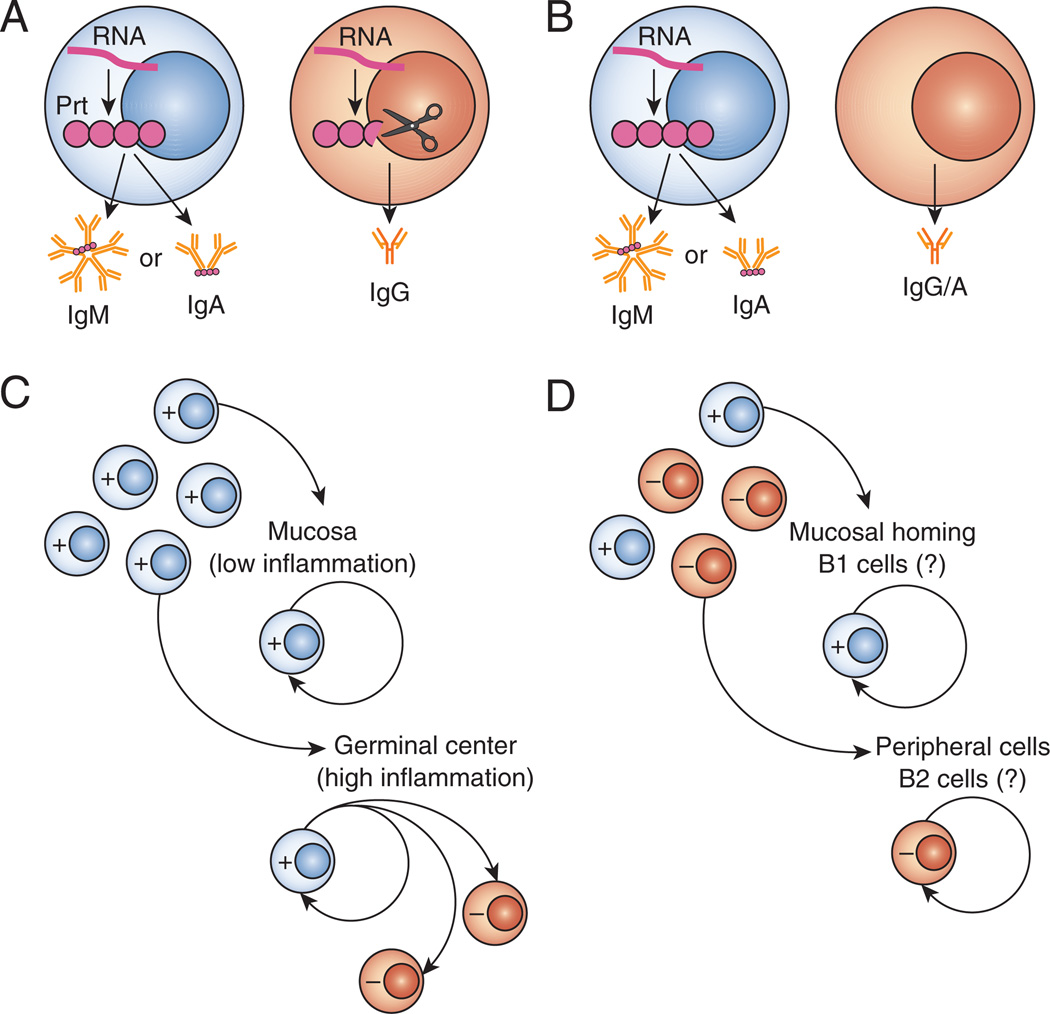
Figure 2. Models of J-chain expression.
(A–B) The presence or absence of J-chain transcript (wavy line) and protein (filled circles) in J-chain+ cells (blue) and J-chain− secretory B cells (pink) are shown. Pentameric IgM and dimeric IgA would be secreted from cells expressing both RNA and protein (blue, A and B). In IgG-secreting cells, J-chain RNA may still be present but J-chain protein is degraded by proteases (scissors) when it does not associate with secretory Ig (pink, A) (65, 67). Alternatively, perhaps neither J-chain RNA nor protein is present in cells secreting monomeric Ig (in this case either IgG or IgA, pink, B) (70). Work from the Brandtzaeg lab suggests that all activated B cells express J-chain by default (blue, C), and only after prolonged clonal proliferation or germinal center reactions do the activated B cells become J-chain-negative (pink, C) (16, 75, 76). Conversely, evidence from other groups suggests that some B cells are marked at an early stage to be either J-chain+ (blue) or J-chain− (pink, D (20)) and that the J-chain+ (blue, D) B cells may be derived from a different lineage than J-chain− cells (90).
To address the former point: if J-chain were transcribed in all IgA-secreting cells, one would assume it would be able to associate with Ig, and therefore all secreted IgA would be dimeric. We therefore believe that J-chain must be repressed in some IgA secreting cells through some Pax5-independent mechanism to allow for secretion of monomeric IgA (Figure 1B, 2B). If J-chain protein is expressed, however, what prevents its association with Ig in monomeric IgA-secreting cells? What signals might direct its decay or otherwise block its association with IgA? Early work suggested that J-chain transcripts were found differentially on free ribosomes or associated with rough ER (RER) and Golgi depending on whether or not secretory Ig was being produced (69), so perhaps some sort of sequestration could occur. These findings were based solely on immuno-EM staining, and since no mechanism of regulation was suggested this idea requires additional support.
As for the latter issue: certain types of IgG-secreting B cells are more likely than others to express J-chain, and specifically, in IgA-deficient humans, IgG1- and IgG2-secreting cells have higher levels of J-chain expression than other IgG subtypes (68). So why is J-chain protein not degraded in all IgG+ cells, if it is degraded in some? An alternative suggestion could be that, like monomeric IgA-secreting cells, it is simply absent in certain cell subsets (Figure 1B, 2B).
Based on these questions it will be important to determine whether J-chain regulation truly occurs at the protein level or if could it be regulated at a more basic level, as in sharks. Some shark plasma cells expressing monomeric IgM lack J-chain RNA, despite having high levels of Blimp1 (Figure 1B) suggesting that there may be other factors present to repress J-chain expression (70) (more detail below).
Populations of J-chain expressing and non-expressing plasma cells
Early work on J-chain suggested it was a feature of B cell activation, where J-chain was initially expressed after activation, and then repressed after prolonged clonal proliferation of antigen-responsive cells (16, 71). These findings were supported by other studies suggesting that J-chain expression is an early event in B cell differentiation (72, 73) and could be a marker of young memory clones (74, 75). However, there are subtle differences between this finding and other early models of J-chain expression, which now should be re-evaluated with existing knowledge of B cell development/activation.
The Brandtzaeg group proposed that J-chain expression is stage-specific. During an ongoing immune response, they found that early plasma cells, present during the height of inflammation, were J-chain+, while later, higher affinity, derived clones did not express J-chain (16, 75, 76) (Figure 2C). In contrast, Erlandsson, et al., using diphtheria toxin A (DTA) targeting to the J-chain locus, found that cells are clonally marked from an early stage of development to be either J-chain-positive or -negative, long before they become Ig-secreting cells (20) (Figure 2D). In humans, J-chain expression has also been described in early B cell development, prior to antigen receptor expression (2, 69, 77, 78), but how this early J-chain expression fits in with “clonally marked” J-chain-positive or -negative cells is unclear.
J-chain in B1 vs. B2 cells
Mammalian B cells are subdivided into three main lineages: B2 (follicular B cells), B1, and Marginal Zone (MZ) B cells, based on developmental appearance, tissue localization, cell surface markers, BCR repertoires, and response to antigen. B1 cells, composed of B1a or B1b cells, are considered to be “innate-like” B cells (79), which differentiate early in development from a distinct B1 cell precursor, express a unique B cell receptor repertoire (80–82), and, as plasma cells, can be induced to secrete “natural antibodies” (83, 84). B1 cells are found in the peritoneal cavity and LP of the intestine, and rarely in secondary lymphoid tissues. This unique, LP-associated, localization of B1 cells marks these cells as important for the production of multimeric, J-chain associated Ig isotypes that are secreted into the lumen. J-chain has been described as a marker of mucosal-targeted plasma cells (Figure 2C, D), wherein the presence of J-chain in some human IgD+ and IgG+ cells is explained by their mucosal-associated location (66).
Dimeric IgA found in mucosal secretions is polyreactive and specific for commensal bacteria (85, 86), and it has been suggested that B1 cells are responsible for production of this commensal-specific IgA (87). B2 cell-derived IgA, in contrast, is found in the serum and has a different V gene repertoire than B1-derived antibodies (88, 89). Therefore, the expression of J-chain may be indicative of B1-derived IgA secreting cells. In support of this, evidence suggests that J-chain−, monomeric IgA secreting cells are derived from either the B1b or B2 cell lineages, while the J-chain+, dimeric IgA-secreting cells arise from cells of the B1a lineage (90). This model of J-chain expression, as an inherent trait of certain B cell lineages, fits well with Leanderson’s idea that J-chain expression is a clonal property (Figure 2D) (20), yet it could also explain why “mucosal-associated” B cells tend to be J-chain+ as proposed by Brandtzaeg (Figure 2C) (66).
There are, however, some conflicting and complicating views on the origins of these J-chain+ IgA-secreting cells. Both IgA and IgM function at mucosal sites, and IgM can compensate for IgA to some degree in the intestinal lumen (e.g. in binding to commensal organisms), explaining why there is not a robust phenotype in IgA-deficient animals. It was reported that in mice expressing the lambda2 L chain, in which all cells are committed to the B1 lineage (91), the majority of IgA is not secreted by B1a cells, and instead B1a cells produce mostly IgM (92). Additionally, it has been shown that AID-dependent mutations are required for control of mucosal bacteria (93). Since B1 cells are generally thought not to undergo high levels of mutation, this suggests that the cells responsible for this commensal control are B2-derived, J-chain-expressing, IgA+ cells, rather than B1-derived cells; however, although the B1 cell BCR repertoire is typically reported to be of low diversity (unmutated, few N-nucleotides), this too is controversial, as it has been suggested that IgH in B1 cells can be highly mutated in some cases (94, 95).
At the moment, results from the experiments in the previous paragraph are difficult to reconcile, however several recent reports suggest that the presence of certain helper T cell types or bacterial species could also have an affect on “innate like” IgA. Specifically, the presence of segmented filamentous bacteria (SFP) has been linked to an increase in IgA production, and this IgA is less mutated and more polyreactive than that produced in response to E. coli, suggesting that it may be more innate-like (96). Additionally, SFB may regulate TH17 cells, and in some cases these TH17 cells may act as progenitors for TFH cells, which can, in turn, support high levels of IgA secretion (97), although eosinophils have also recently been implicated in high levels of T-independent IgA secretion (98). These recent studies suggest that the local mucosal environment play a role in secretion of “innate-like” IgA that is more nuanced than simply the presence of B1 vs. B2 derived cells. It is important to note that there was no analysis of J-chain expression and/or the multimerization state of IgA in these recent reports, so it is not clear if the J-chain expression is more similar to the model outlined in Figure 2C or 2D.
As mentioned previously, additional complications arise when comparing human and mouse IgA. Unlike mice, humans have 2 IgA isotypes, IgA1 and IgA2, and the latter preferentially associates with J-chain (50), which could partially explain how regulation of monomeric IgA secretion is achieved. However, this model is not feasible in mice, which only possess one IgA gene. Little is known about regulation of J-chain expression in these cells, but further study would be quite informative, i.e. factors regulating the class switch might also regulate J-chain expression (99). Our group has recently shown that, in nurse sharks, the control of J-chain in plasma cells must occur at the expression level, as J-chain RNA transcripts are only found in a subset of plasma cells. In contrast to the prevailing transcriptional model in mammals that Blimp1 relieves Pax5 repression for J-chain expression (Figure 1A), some shark plasma cells are Blimp1+ but do NOT express J-chain (Figure 1B). Is the same true of B cell subsets in mammals, and how does that help us to understand J-chain regulation? We have also found that these cells have other unusual expression patterns for plasma cells, specifically that J-chain+ cells in the nurse shark are Blimp1-negative (Figure 1C) (70). It would therefore be interesting to examine the levels of J-chain and Blimp1 RNA in mouse (or human) monomeric IgA-secreting cells, and to determine which other transcription factors may be playing a role in this regulation. In the case of nurse shark plasma cells, the observed J-chain expression pattern could fit with either discussed model: expression in early plasmablasts, which is extinguished late in activation (Figure 2C) or expression in dedicated B cell subsets (Figure 2D).
Conclusions
Many mysteries persist in the study of J-chain, and not all current proposed discussions take into account how regulation of this molecule/gene is controlled. For one: seemingly, the J-chain ‘came out of nowhere’ during early evolution of the adaptive immune system (1); which gene family gave rise to the J-chain, and how was it co-opted by the immune system? We anxiously await the first crystal structure of an IgM molecule, which would finally reveal J-chain’s secrets (as well as those of IgM!). Besides B cells, J-chain expression has been detected in developing lymphocytes (30), DCs (32), and intestinal epithelial cells (34), none of which express secretory forms of Ig heavy chains. Do these studies point to novel, perhaps primordial, functions for J-chain? Conversely, is the unusual J-chain expression simply a function of the combination of transcription factors expressed in certain cells?
How does mucosal localization of plasma cell subsets affect their J-chain expression profile? In the mucosal LP, J-chain expression is promoted in some cells under the influence of unknown signals and transcription factors. Could these signals be coming from particular T-helper subsets, stroma or myeloid cells, or perhaps from the same eosinophils and/or TH17 or TFH cells responsible for extrafolicular switch to IgA? Since inflammation is generally suppressed in the intestinal epithelium, does this control somehow contribute to upregulation of J-chain? What is the contribution of T-independent class switch recombination (CSR) in LP to J-chain expression, and might it be related to studies suggesting that J-chain is found in secretory cells expressing only certain IgG isotypes (68)? Studies of non-mammalian vertebrates (amphibians, bony fish, and sharks) described above (37–40, 70) could provide definitive models for studying the question of J-chain expression in the mucosa. For example, we would predict that IgX-producing frog cells found in the LP would express J-chain because of their mucosal localization (38), even though the H chain not associate with this isotype due to a lack of the canonical cysteine in the secretory tail (37).
Perhaps most importantly, it is still unclear which exact factors affect J-chain expression in plasma cells. Current understanding of J-chain regulation is based on similarities to the Igκ promoter and relief of Pax5 repression (11, 64), but different models of J-chain expression in plasma cells propose either an ordered expression of J-chain during an immune response in plasma cells (16, 75, 76) or expression in different subsets of B cells, clonally marked from early development (20) (Figure 2). Future studies of J-chain gene control, would be quite useful in not only examining J-chain expression but also the general gene expression programs of plasma cell subsets. With the recognized importance of IgA to mucosal homeostasis and barrier function, and the significance of J-chain for proper IgA multimerization and transport, we believe that understanding the true nature of J-chain regulation has important implications for human health and disease, particularly with the current rise in intestinal disorders.
Footnotes
This work was supported by Grant R01OD0549 from the National Institutes of Health as well as Institutional Training Grants in Cardiac and Vascular Cell Biology (5T32HL072751) and Immunity and Infection (T32AI007540) from The National Institute of Allergy and Infection Diseases
References
- 1.Hohman VS, Stewart SE, Rumfelt LL, Greenberg AS, Avila DW, Flajnik MF, Steiner LA. J chain in the nurse shark: implications for function in a lower vertebrate. J. Immunol. 2003;170:6016–6023. doi: 10.4049/jimmunol.170.12.6016. [DOI] [PubMed] [Google Scholar]
- 2.Max EE, Korsmeyer SJ. Human J chain gene. Structure and expression in B lymphoid cells. J. Exp. Med. 1985;161:832–849. doi: 10.1084/jem.161.4.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastian A, Kratzin H, Fallgren-Gebauer E, Eckart K, Hilschmann N. Intra- and inter-chain disulfide bridges of J chain in human S-IgA. Adv. Exp. Med. Biol. 1995;371A:581–583. doi: 10.1007/978-1-4615-1941-6_122. [DOI] [PubMed] [Google Scholar]
- 4.Kallberg E, Leandersson T. Analysis of antigen-specific and naturally occurring IgM and IgA steady-state levels in J-chain negative C57BL/6 mice. Scand. J. Immunol. 2006;63:430–434. doi: 10.1111/j.1365-3083.2006.001762.x. [DOI] [PubMed] [Google Scholar]
- 5.Brandtzaeg P. Immunohistochemical characterization of intracellular J-chain and binding site for secretory component (SC) in human immunoglobulin (Ig)-producing cells. Mol. Immunol. 1983;20:941–966. doi: 10.1016/0161-5890(83)90036-6. [DOI] [PubMed] [Google Scholar]
- 6.Mestecky J, McGhee JR. Immunoglobulin A (IgA): molecular and cellular interactions involved in IgA biosynthesis and immune response. Adv. Immunol. 1987;40:153–245. doi: 10.1016/s0065-2776(08)60240-0. [DOI] [PubMed] [Google Scholar]
- 7.Johansen FE, Braathen R, Brandtzaeg P. Role of J chain in secretory immunoglobulin formation. Scand. J. Immunol. 2000;52:240–248. doi: 10.1046/j.1365-3083.2000.00790.x. [DOI] [PubMed] [Google Scholar]
- 8.Hendrickson BA, Conner DA, Ladd DJ, Kendall D, Casanova JE, Corthesy B, Max EE, Neutra MR, Seidman CE, Seidman JG. Altered hepatic transport of immunoglobulin A in mice lacking the J chain. J. Exp. Med. 1995;182:1905–1911. doi: 10.1084/jem.182.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandtzaeg P. Mucosal immunity: induction, dissemination, and effector functions. Scand. J. Immunol. 2009;70:505–515. doi: 10.1111/j.1365-3083.2009.02319.x. [DOI] [PubMed] [Google Scholar]
- 10.Brandtzaeg P, Prydz H. Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature. 1984;311:71–73. doi: 10.1038/311071a0. [DOI] [PubMed] [Google Scholar]
- 11.Rinkenberger JL, Wallin JJ, Johnson KW, Koshland ME. An interleukin-2 signal relieves BSAP (Pax5)-mediated repression of the immunoglobulin J chain gene. Immunity. 1996;5:377–386. doi: 10.1016/s1074-7613(00)80263-0. [DOI] [PubMed] [Google Scholar]
- 12.Rao S, Karray S, Gackstetter ER, Koshland ME. Myocyte enhancer factor-related B-MEF2 is developmentally expressed in B cells and regulates the immunoglobulin J chain promoter. J. Biol. Chem. 1998;273:26123–26129. doi: 10.1074/jbc.273.40.26123. [DOI] [PubMed] [Google Scholar]
- 13.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell. Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crago SS, Kutteh WH, Moro I, Allansmith MR, Radl J, Haaijman JJ, Mestecky J. Distribution of IgA1-, IgA2-, and J chain-containing cells in human tissues. J. Immunol. 1984;132:16–18. [PubMed] [Google Scholar]
- 16.Brandtzaeg P, Korsrud FR. Significance of different J chain profiles in human tissues: generation of IgA and IgM with binding site for secretory component is related to the J chain expressing capacity of the total local immunocyte population, including IgG and IgD producing cells, and depends on the clinical state of the tissue. Clin. Exp. Immunol. 1984;58:709–718. [PMC free article] [PubMed] [Google Scholar]
- 17.Koshland ME. The coming of age of the immunoglobulin J chain. Annu. Rev. Immunol. 1985;3:425–453. doi: 10.1146/annurev.iy.03.040185.002233. [DOI] [PubMed] [Google Scholar]
- 18.Mestecky J, Zikan J, Butler WT. Immunoglobulin M and secretory immunoglobulin A: presence of a common polypeptide chain different from light chains. Science. 1971;171:1163–1165. doi: 10.1126/science.171.3976.1163. [DOI] [PubMed] [Google Scholar]
- 19.Erlandsson L, Andersson K, Sigvardsson M, Lycke N, Leanderson T. Mice with an inactivated joining chain locus have perturbed IgM secretion. Eur. J. Immunol. 1998;28:2355–2365. doi: 10.1002/(SICI)1521-4141(199808)28:08<2355::AID-IMMU2355>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 20.Erlandsson L, Akerblad P, Vingsbo-Lundberg C, Kallberg E, Lycke N, Leanderson T. Joining chain-expressing and -nonexpressing B cell populations in the mouse. J. Exp. Med. 2001;194:557–570. doi: 10.1084/jem.194.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klimovich VB, Samoilovich MP, Klimovich BV. Problem of J-chain of immunoglobulins. Zh. Evol. Biokhim. Fiziol. 2008;44:131–143. [PubMed] [Google Scholar]
- 22.Cattaneo A, Neuberger MS. Polymeric immunoglobulin M is secreted by transfectants of non-lymphoid cells in the absence of immunoglobulin J chain. EMBO J. 1987;6:2753–2758. doi: 10.1002/j.1460-2075.1987.tb02569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randall TD, Brewer JW, Corley RB. Direct evidence that J chain regulates the polymeric structure of IgM in antibody-secreting B cells. J. Biol. Chem. 1992;267:18002–18007. [PubMed] [Google Scholar]
- 24.Davis AC, Roux KH, Shulman MJ. On the structure of polymeric IgM. Eur. J. Immunol. 1988;18:1001–1008. doi: 10.1002/eji.1830180705. [DOI] [PubMed] [Google Scholar]
- 25.Brewer JW, Randall TD, Parkhouse RM, Corley RB. Mechanism and subcellular localization of secretory IgM polymer assembly. J. Biol. Chem. 1994;269:17338–17348. [PubMed] [Google Scholar]
- 26.Kownatzki E. Reassociation of IgM subunits in the presence and absence of J chain. Immunol. Commun. 1973;2:105–113. doi: 10.3109/08820137309022886. [DOI] [PubMed] [Google Scholar]
- 27.Eskeland T, Christensen TB. IgM molecules with and without J chain in serum and after purification, studied by ultracentrifugation, electrophoresis, and electron microscopy. Scand. J. Immunol. 1975;4:217–228. doi: 10.1111/j.1365-3083.1975.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 28.Petrusic V, Zivkovic I, Stojanovic M, Stojicevic I, Marinkovic E, Dimitrijevic L. Hexameric immunoglobulin M in humans: desired or unwanted? Med. Hypotheses. 2011;77:959–961. doi: 10.1016/j.mehy.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Petrusic V, Stojanovic M, Zivkovic I, Inic-Kanada A, Dimitrijevic L. Changes in composition of IgM polymers in patients suffering from recurrent urinary bacterial infections after bacterial immunization treatment. Immunol. Invest. 2010;39:781–795. doi: 10.3109/08820139.2010.497831. [DOI] [PubMed] [Google Scholar]
- 30.Bertrand FE, 3rd, Billips LG, Gartland GL, Kubagawa H, Schroeder HW., Jr The J chain gene is transcribed during B and T lymphopoiesis in humans. J. Immunol. 1996;156:4240–4244. [PubMed] [Google Scholar]
- 31.Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint Vis B, Briere F, Bates EE. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 32.Kallberg E, Leanderson T. A subset of dendritic cells express joining chain (J-chain) protein. Immunology. 2008;123:590–599. doi: 10.1111/j.1365-2567.2007.02733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kallberg E, Leanderson T. Joining-chain (J-chain) negative mice are B cell memory deficient. Eur. J. Immunol. 2006;36:1398–1403. doi: 10.1002/eji.200635981. [DOI] [PubMed] [Google Scholar]
- 34.Tacchi L, Larragoite E, Salinas I. Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: implications in mucosal immunity. PLoS One. 2013;8:e70650. doi: 10.1371/journal.pone.0070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat. Rev. Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- 36.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mussmann R, Du Pasquier L, Hsu E. Is Xenopus IgX an analog of IgA? Eur. J. Immunol. 1996;26:2823–2830. doi: 10.1002/eji.1830261205. [DOI] [PubMed] [Google Scholar]
- 38.Mashoof S, Goodroe A, Du CC, Eubanks JO, Jacobs N, Steiner JM, Tizard I, Suchodolski JS, Criscitiello MF. Ancient T-independence of mucosal IgX/A: gut microbiota unaffected by larval thymectomy in Xenopus laevis. Mucosal Immunol. 2013;6:358–368. doi: 10.1038/mi.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flajnik MF. All GOD's creatures got dedicated mucosal immunity. Nat. Immunol. 2010;11:777–779. doi: 10.1038/ni0910-777. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi T, Iwase T, Takenouchi N, Saito M, Kobayashi K, Moldoveanu Z, Mestecky J, Moro I. The joining (J) chain is present in invertebrates that do not express immunoglobulins. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1886–1891. doi: 10.1073/pnas.93.5.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamuro K, Suetake H, Saha NR, Kikuchi K, Suzuki Y. A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J. Immunol. 2007;178:5682–5689. doi: 10.4049/jimmunol.178.9.5682. [DOI] [PubMed] [Google Scholar]
- 43.Feng LN, Lu DQ, Bei JX, Chen JL, Liu Y, Zhang Y, Liu XC, Meng ZN, Wang L, Lin HR. Molecular cloning and functional analysis of polymeric immunoglobulin receptor gene in orange-spotted grouper (Epinephelus coioides) Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2009;154:282–289. doi: 10.1016/j.cbpb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Kortum AN, Rodriguez-Nunez I, Yang J, Shim J, Runft D, O'Driscoll ML, Haire RN, Cannon JP, Turner PM, Litman RT, Kim CH, Neely MN, Litman GW, Yoder JA. Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics. 2014 doi: 10.1007/s00251-014-0759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]
- 46.Hsu E, Flajnik MF, Du Pasquier L. A third immunoglobulin class in amphibians. J. Immunol. 1985;135:1998–2004. [PubMed] [Google Scholar]
- 47.Anderson MK, Pant R, Miracle AL, Sun X, Luer CA, Walsh CJ, Telfer JC, Litman GW, Rothenberg EV. Evolutionary origins of lymphocytes: ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J. Immunol. 2004;172:5851–5860. doi: 10.4049/jimmunol.172.10.5851. [DOI] [PubMed] [Google Scholar]
- 48.Zwollo P. Dissecting teleost B cell differentiation using transcription factors. Dev. Comp. Immunol. 2011;35:898–905. doi: 10.1016/j.dci.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burnett RC, Hanly WC, Zhai SK, Knight KL. The IgA heavy-chain gene family in rabbit: cloning and sequence analysis of 13 C alpha genes. EMBO J. 1989;8:4041–4047. doi: 10.1002/j.1460-2075.1989.tb08587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bjerke K, Brandtzaeg P. Terminally differentiated human intestinal B cells. J chain expression of IgA and IgG subclass-producing immunocytes in the distal ileum compared with mesenteric and peripheral lymph nodes. Clin. Exp. Immunol. 1990;82:411–415. doi: 10.1111/j.1365-2249.1990.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, Knowles DM, Rescigno M, Cerutti A. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Marchalonis J, Edelman GM. Phylogenetic origins of antibody structure. I. Multichain structure of immunoglobulins in the smooth dogfish (Mustelus canis) J. Exp. Med. 1965;122:601–618. doi: 10.1084/jem.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clem IW, De Boutaud F, Sigel MM. Phylogeny of immunoglobulin structure and function. II. Immunoglobulins of the nurse shark. J. Immunol. 1967;99:1226–1235. [PubMed] [Google Scholar]
- 54.Frommel D, Litman GW, Finstad J, Good RA. The evolution of the immune response. XI. The immunoglobulins of the horned shark, Heterodontus francisci: purification, characterization and structural requirement for antibody activity. J. Immunol. 1971;106:1234–1243. [PubMed] [Google Scholar]
- 55.McCumber LJ, Clem LW. A comparative study of J chain structure and stoichiometry in human and nurse shark IgM. Immunochemistry. 1976;13:479–484. doi: 10.1016/0019-2791(76)90322-0. [DOI] [PubMed] [Google Scholar]
- 56.Klapper DG, Clem LW. Phylogeny of immunoglobulin structure and function; characterization of the cysteine-containing peptide involved in the pentamerization of shark IgM. Dev. Comp. Immunol. 1977;1:81–91. doi: 10.1016/s0145-305x(77)80002-5. [DOI] [PubMed] [Google Scholar]
- 57.Nutt SL, Eberhard D, Horcher M, Rolink AG, Busslinger M. Pax5 determines the identity of B cells from the beginning to the end of B-lymphopoiesis. Int. Rev. Immunol. 2001;20:65–82. doi: 10.3109/08830180109056723. [DOI] [PubMed] [Google Scholar]
- 58.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat. Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 59.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 60.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 61.Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J. Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 62.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 64.Sigvardsson M, Olsson L, Hogbom E, Leanderson T. Characterization of the joining chain (J-chain) promoter. Scand. J. Immunol. 1993;38:411–416. doi: 10.1111/j.1365-3083.1993.tb02581.x. [DOI] [PubMed] [Google Scholar]
- 65.Mosmann TR, Gravel Y, Williamson AR, Baumal R. Modification and fate of J chain in myeloma cells in the presence and absence of polymeric immunoglobulin secretion. Eur. J. Immunol. 1978;8:94–101. doi: 10.1002/eji.1830080205. [DOI] [PubMed] [Google Scholar]
- 66.Brandtzaeg P, Johansen FE. Mucosal B cells: phenotypic characteristics, transcriptional regulation, and homing properties. Immunol. Rev. 2005;206:32–63. doi: 10.1111/j.0105-2896.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 67.Lamson G, Koshland ME. Changes in J chain and mu chain RNA expression as a function of B cell differentiation. J. Exp. Med. 1984;160:877–892. doi: 10.1084/jem.160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilssen DE, Brandtzaeg P, Froland SS, Fausa O. Subclass composition and J-chain expression of the 'compensatory' gastrointestinal IgG cell population in selective IgA deficiency. Clin. Exp. Immunol. 1992;87:237–245. doi: 10.1111/j.1365-2249.1992.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hajdu I, Moldoveanu Z, Cooper MD, Mestecky J. Ultrastructural studies of human lymphoid cells. mu and J chain expression as a function of B cell differentiation. J. Exp. Med. 1983;158:1993–2006. doi: 10.1084/jem.158.6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castro CD, Ohta Y, Dooley H, Flajnik MF. Noncoordinate expression of J-chain and Blimp-1 define nurse shark plasma cell populations during ontogeny. Eur. J. Immunol. 2013;43:3061–3075. doi: 10.1002/eji.201343416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brandtzaeg P. Presence of J chain in human immunocytes containing various immunoglobulin classes. Nature. 1974;252:418–420. doi: 10.1038/252418a0. [DOI] [PubMed] [Google Scholar]
- 72.Koshland ME. Presidential address: molecular aspects of B cell differentiation. American Association of Immunologists April 1983. J. Immunol. 1983;131:i–ix. [PubMed] [Google Scholar]
- 73.Kutteh WH, Moldoveanu Z, Prince SJ, Kulhavy R, Alonso F, Mestecky J. Biosynthesis of J-chain in human lymphoid cells producing immunoglobulins of various isotypes. Mol. Immunol. 1983;20:967–976. doi: 10.1016/0161-5890(83)90037-8. [DOI] [PubMed] [Google Scholar]
- 74.Brandtzaeg P. Studies on J chain and binding site for secretory component in circulating human B cells. II. The cytoplasm. Clin. Exp. Immunol. 1976;25:59–66. [PMC free article] [PubMed] [Google Scholar]
- 75.Korsrud FR, Brandtzaeg P. Immunohistochemical evaluation of J-chain expression by intra- and extra-follicular immunoglobulin-producing human tonsillar cells. Scand. J. Immunol. 1981;13:271–280. doi: 10.1111/j.1365-3083.1981.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 76.Kett K, Brandtzaeg P, Fausa O. J-chain expression is more prominent in immunoglobulin A2 than in immunoglobulin A1 colonic immunocytes and is decreased in both subclasses associated with inflammatory bowel disease. Gastroenterology. 1988;94:1419–1425. doi: 10.1016/0016-5085(88)90681-6. [DOI] [PubMed] [Google Scholar]
- 77.McCune JM, Fu SM, Kunkel HG. J chain biosynthesis in pre-B cells and other possible precursor B cells. J. Exp. Med. 1981;154:138–145. doi: 10.1084/jem.154.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kubagawa H, Burrows PD, Grossi CE, Mestecky J, Cooper MD. Precursor B cells transformed by Epstein-Barr virus undergo sterile plasma-cell differentiation: J-chain expression without immunoglobulin. Proc. Natl. Acad. Sci. U. S. A. 1988;85:875–879. doi: 10.1073/pnas.85.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baumgarth N, Herman OC, Jager GC, Brown L, Herzenberg LA, Herzenberg LA. Innate and acquired humoral immunities to influenza virus are mediated by distinct arms of the immune system. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2250–2255. doi: 10.1073/pnas.96.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kantor AB, Herzenberg LA. Origin of murine B cell lineages. Annu. Rev. Immunol. 1993;11:501–538. doi: 10.1146/annurev.iy.11.040193.002441. [DOI] [PubMed] [Google Scholar]
- 81.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat. Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 82.Herzenberg LA, Tung JW. B cell lineages: documented at last! Nat. Immunol. 2006;7:225–226. doi: 10.1038/ni0306-225. [DOI] [PubMed] [Google Scholar]
- 83.Thurnheer MC, Zuercher AW, Cebra JJ, Bos NA. B1 cells contribute to serum IgM, but not to intestinal IgA, production in gnotobiotic Ig allotype chimeric mice. J. Immunol. 2003;170:4564–4571. doi: 10.4049/jimmunol.170.9.4564. [DOI] [PubMed] [Google Scholar]
- 84.Choi YS, Dieter JA, Rothaeusler K, Luo Z, Baumgarth N. B-1 cells in the bone marrow are a significant source of natural IgM. Eur. J. Immunol. 2012;42:120–129. doi: 10.1002/eji.201141890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quan CP, Berneman A, Pires R, Avrameas S, Bouvet JP. Natural polyreactive secretory immunoglobulin A autoantibodies as a possible barrier to infection in humans. Infect. Immun. 1997;65:3997–4004. doi: 10.1128/iai.65.10.3997-4004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shimoda M, Inoue Y, Azuma N, Kanno C. Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer's patches. Immunology. 1999;97:9–17. doi: 10.1046/j.1365-2567.1999.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kroese FG, Butcher EC, Stall AM, Lalor PA, Adams S, Herzenberg LA. Many of the IgA producing plasma cells in murine gut are derived from self-replenishing precursors in the peritoneal cavity. Int. Immunol. 1989;1:75–84. doi: 10.1093/intimm/1.1.75. [DOI] [PubMed] [Google Scholar]
- 88.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 89.Stoel M, Jiang HQ, van Diemen CC, Bun JC, Dammers PM, Thurnheer MC, Kroese FG, Cebra JJ, Bos NA. Restricted IgA repertoire in both B-1 and B-2 cell-derived gut plasmablasts. J. Immunol. 2005;174:1046–1054. doi: 10.4049/jimmunol.174.2.1046. [DOI] [PubMed] [Google Scholar]
- 90.Rosado MM, Aranburu A, Capolunghi F, Giorda E, Cascioli S, Cenci F, Petrini S, Miller E, Leanderson T, Bottazzo GF, Natali PG, Carsetti R. From the fetal liver to spleen and gut: the highway to natural antibody. Mucosal Immunol. 2009;2:351–361. doi: 10.1038/mi.2009.15. [DOI] [PubMed] [Google Scholar]
- 91.Engel H, Bogen B, Muller U, Andersson J, Rolink A, Weiss S. Expression level of a transgenic lambda2 chain results in isotype exclusion and commitment to B1 cells. Eur. J. Immunol. 1998;28:2289–2299. doi: 10.1002/(SICI)1521-4141(199808)28:08<2289::AID-IMMU2289>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 92.Roy B, Agarwal S, Brennecke AM, Krey M, Pabst O, Duber S, Weiss S. B-1 cell subpopulations contribute differently to gut immunity. Eur. J. Immunol. 2013 doi: 10.1002/eji.201243070. [DOI] [PubMed] [Google Scholar]
- 93.Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 94.Roy B, Shukla S, Lyszkiewicz M, Krey M, Viegas N, Duber S, Weiss S. Somatic hypermutation in peritoneal B1b cells. Mol. Immunol. 2009;46:1613–1619. doi: 10.1016/j.molimm.2009.02.026. [DOI] [PubMed] [Google Scholar]
- 95.Bos NA, Bun JC, Popma SH, Cebra ER, Deenen GJ, van der Cammen MJ, Kroese FG, Cebra JJ. Monoclonal immunoglobulin A derived from peritoneal B cells is encoded by both germ line and somatically mutated VH genes and is reactive with commensal bacteria. Infect. Immun. 1996;64:616–623. doi: 10.1128/iai.64.2.616-623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, Reynaud CA, Cerf-Bensussan N, Gaboriau-Routhiau V. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 97.Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chu VT, Beller A, Rausch S, Strandmark J, Zanker M, Arbach O, Kruglov A, Berek C. Eosinophils promote generation and maintenance of immunoglobulin-a-expressing plasma cells and contribute to gut immune homeostasis. Immunity. 2014;40:582–593. doi: 10.1016/j.immuni.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 99.Mora JR, von Andrian UH. Differentiation and homing of IgA-secreting cells. Mucosal Immunol. 2008;1:96–109. doi: 10.1038/mi.2007.14. [DOI] [PubMed] [Google Scholar]
- 100.Tumang JR, Frances R, Yeo SG, Rothstein TL. Spontaneously Ig-secreting B-1 cells violate the accepted paradigm for expression of differentiation-associated transcription factors. J. Immunol. 2005;174:3173–3177. doi: 10.4049/jimmunol.174.6.3173. [DOI] [PubMed] [Google Scholar]