Figure 4.
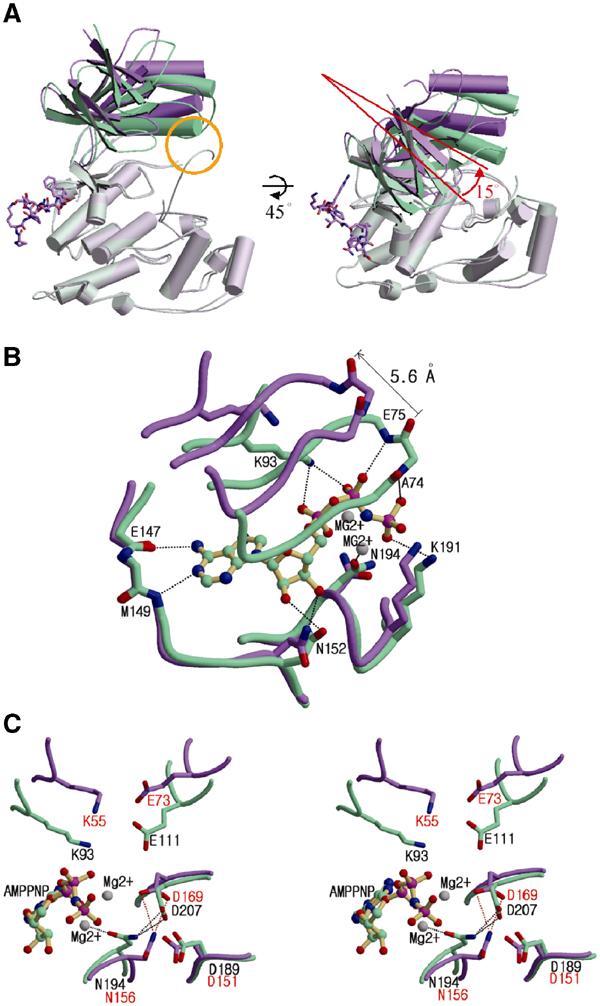
Distortion of the ATP-binding site caused by interdomain rearrangement upon pepJIP1 binding. (A) Structural comparison between JNK3 (green) and pepJIP1-bound JNK1 (violet) when the C-terminal domains of the kinases are superimposed. The conformational differences of the N-terminal domains can be easily distinguished when the conventional view of kinases is rotated by 45° along the horizontal axis. The yellow circle indicates the interaction between the α1 helix and the phosphorylation loop in JNK3, but not existing in JNK1 complexed with pepJIP1. (B) Comparison of ATP-binding sites between the JNK1–pepJIP1 (violet) and JNK3–AMPPNP (green) complexes. The AMPPNP bound in JNK3 is shown in a ball-and-stick model. The residues of JNK3 involved in the hydrogen bonding with AMPPNP are labeled. The side chains of the residues in the glycine-rich loop including E75 and A74 of JNK3 are omitted for clarity because the backbone amide groups only are involved in the hydrogen bonds with the phosphate groups of AMPPNP. (C) The structural comparison of the residues crucial for the catalytic activity between the JNK1–pepJIP1 (violet) and JNK3–AMPPNP (green) complexes. The residues in JNK1 and JNK3 are labeled red and black, respectively. In (B, C), hydrogen bonds are indicated by dashed lines.
