Figure 7.
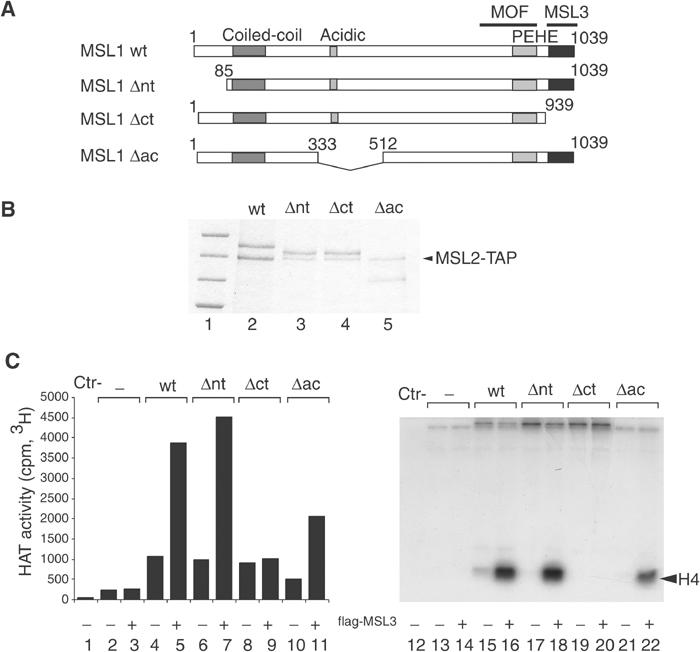
MSL3 binding to MSL1 is required for MOF activation on nucleosomes. (A) Schematic representation of MSL1: wild type (wt) and deletion mutants (Δnt, Δct and Δac). Deleted amino acids are numbered. Characteristic domains of MSL1, like the coiled-coil, the acidic and PEHE domains are indicated. (B) SF9 cells were coinfected with baculoviruses expressing TAP-MSL2 and either wild-type (wt) MSL1 or the MSL1 mutants described in (A). The MSL1 derivatives were purified via the TAP tag of MSL2, and analysed by SDS–PAGE and Coomassie blue staining (lanes 2–5). Lane 1 displays protein size standards in kDa (200, 150, 120, 100, respectively). (C) HAT assays on nucleosome templates were performed in the presence of MOF with or without MSL1 derivatives purified in (B) and in the absence or the presence of purified flag-MSL3. As in Figure 6, the left panel represents the general HAT activity and the right panel the substrate specificity.
