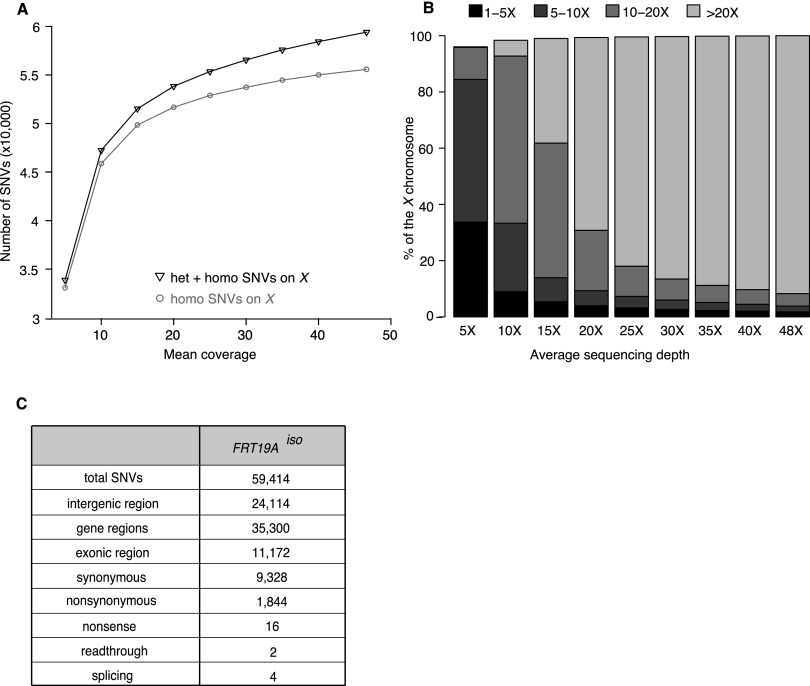
Figure 1.
A sequencing depth of 30× permits identification of 95% of SNVs. (A) Graph displaying the number of identified SNVs at different sequencing depths for the isogenized FRT19A X chromosome (FRT19Aiso). A 30× coverage allows identification of ∼95% of the SNVs identified at 50× coverage. (B) Percentage of the X chromosome that is covered 1×–5× (black), 5×–10× (dark gray), 10×–20× (gray), or ≥20× (light gray) at various average sequencing depths. An average sequencing depth of 30× allows reliable heterozygous SNV-calling (requiring 10 or more reads) of 95% of the X chromosome. (C) Description of SNVs identified in the X chromosome of FRT19Aiso sequenced at 48× when compared to the reference sequence (y; cn bw sp) (Adams et al. 2000).