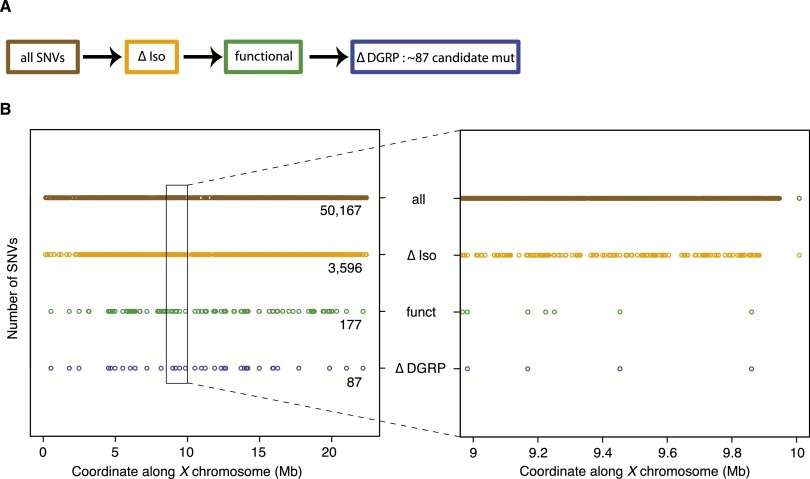
Figure 2.
Filtering process to identify candidate genes in heterozygous mutants. (A) Flowchart of filters applied to identify candidate mutations in heterozygous mutants [y w (*) FRT19A/y w FRT19Aiso]. All identified SNVs (brown) were first filtered against SNVs identified in the isogenized FRT19A X chromosome (ΔIso, orange). Subsequently, only SNVs that affect the coding sequence or splice sites were retained (functional, green). Next, the remaining SNVs were filtered against a database, containing polymorphisms found in a homozygous state in a collection of 205 viable, wild-type strains from the Drosophila Genetic Reference Panel (ΔDGRP, blue). (B, left) Impact of filters, introduced in A, on the total number of SNVs identified on the X chromosome. (Right) In a 1-Mb interval, the number of remaining candidate mutations is ∼4.